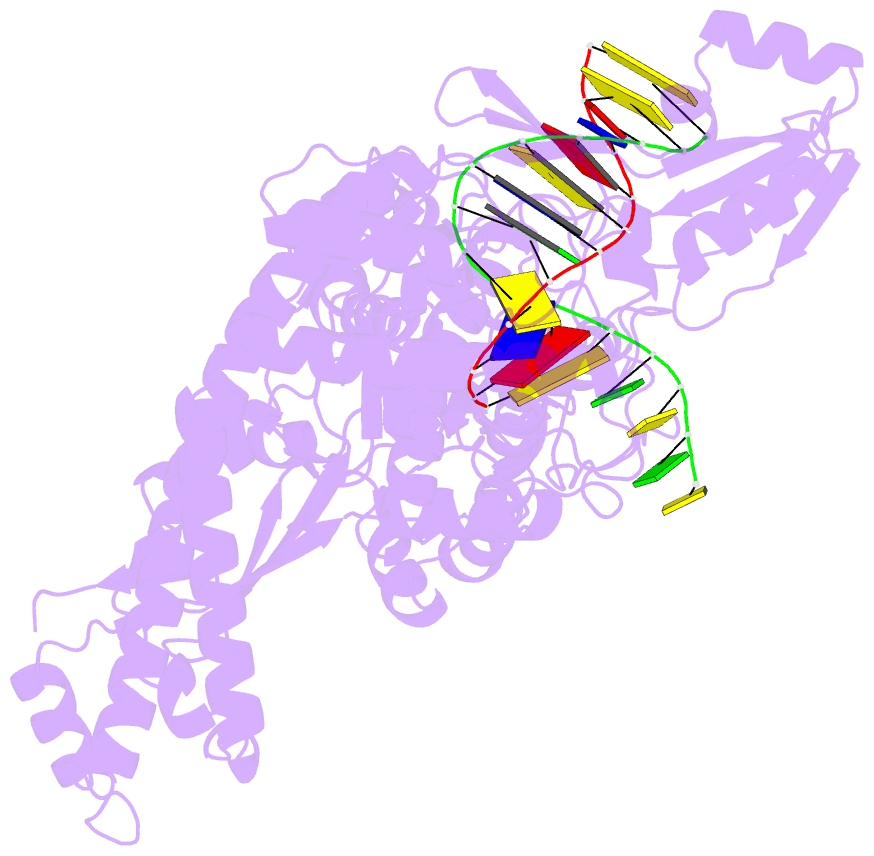
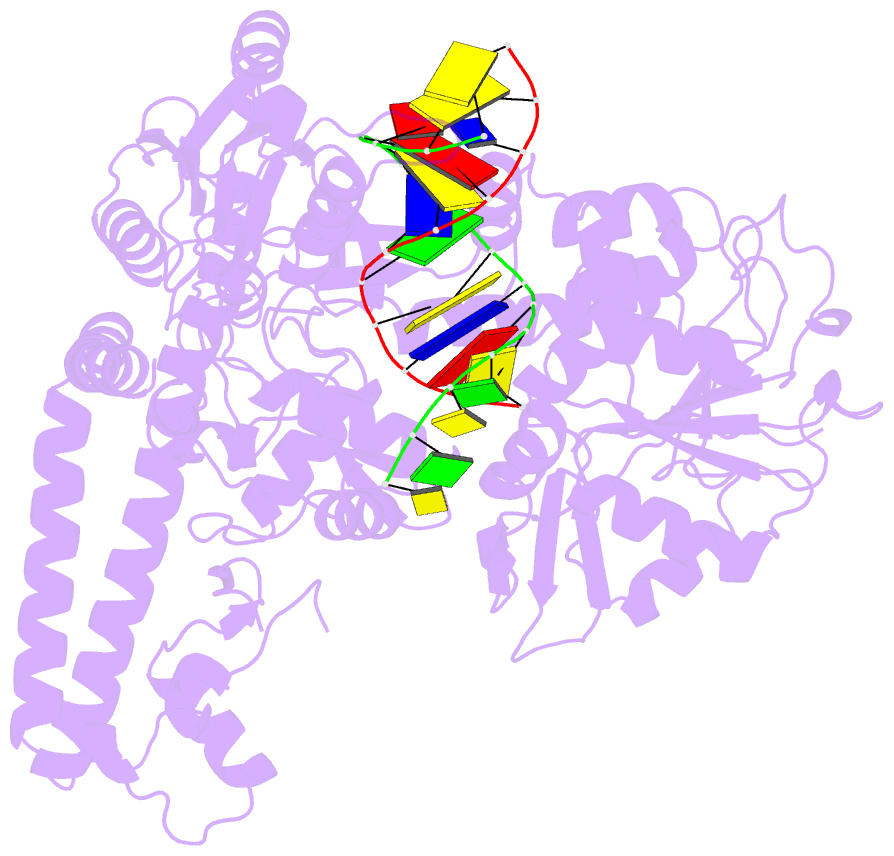
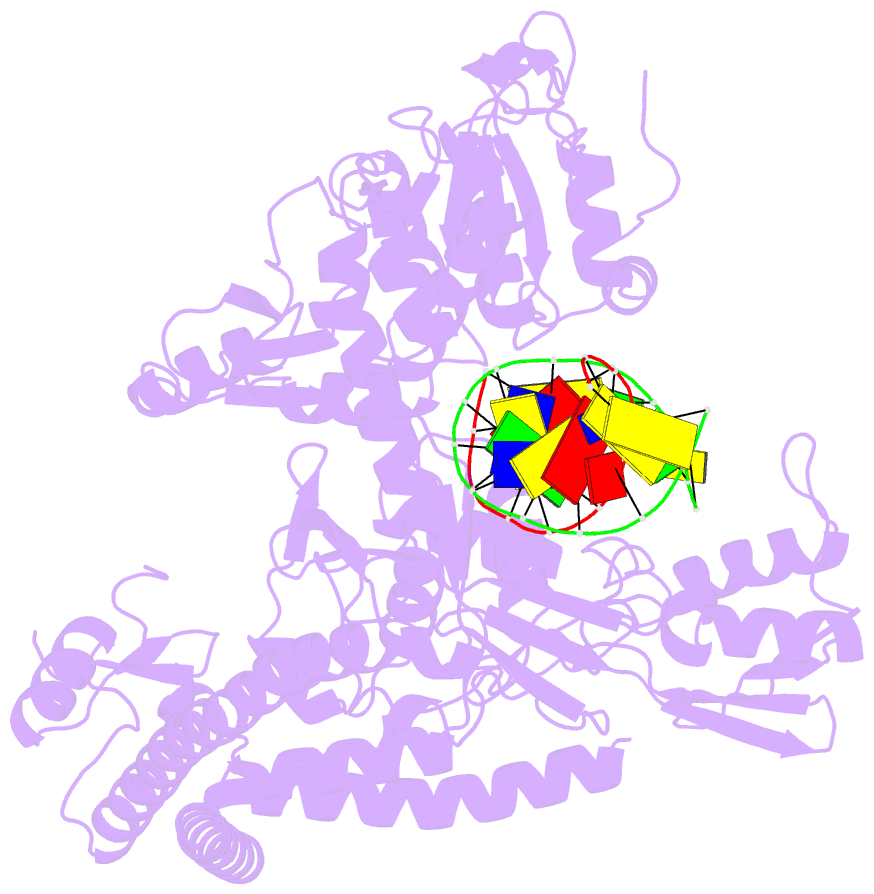
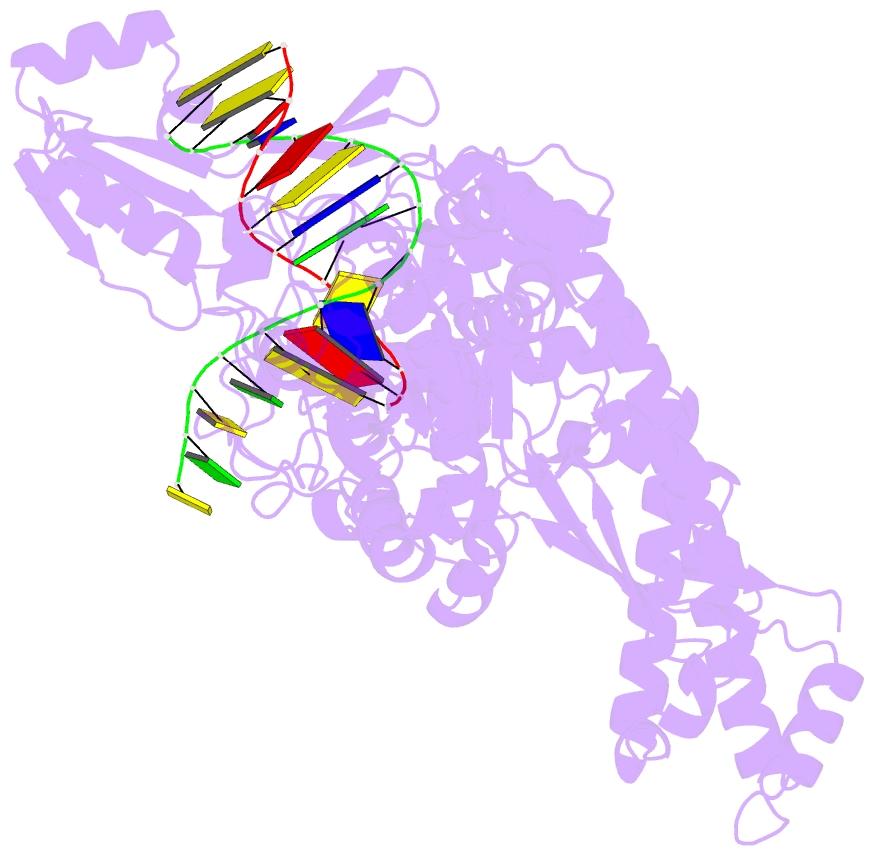
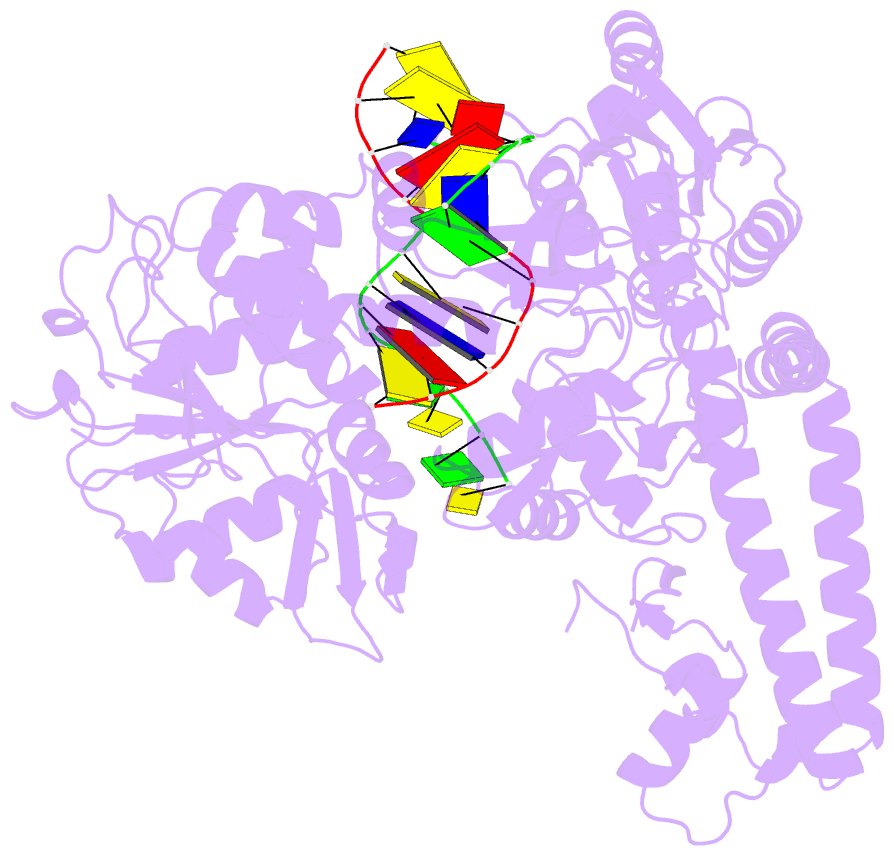
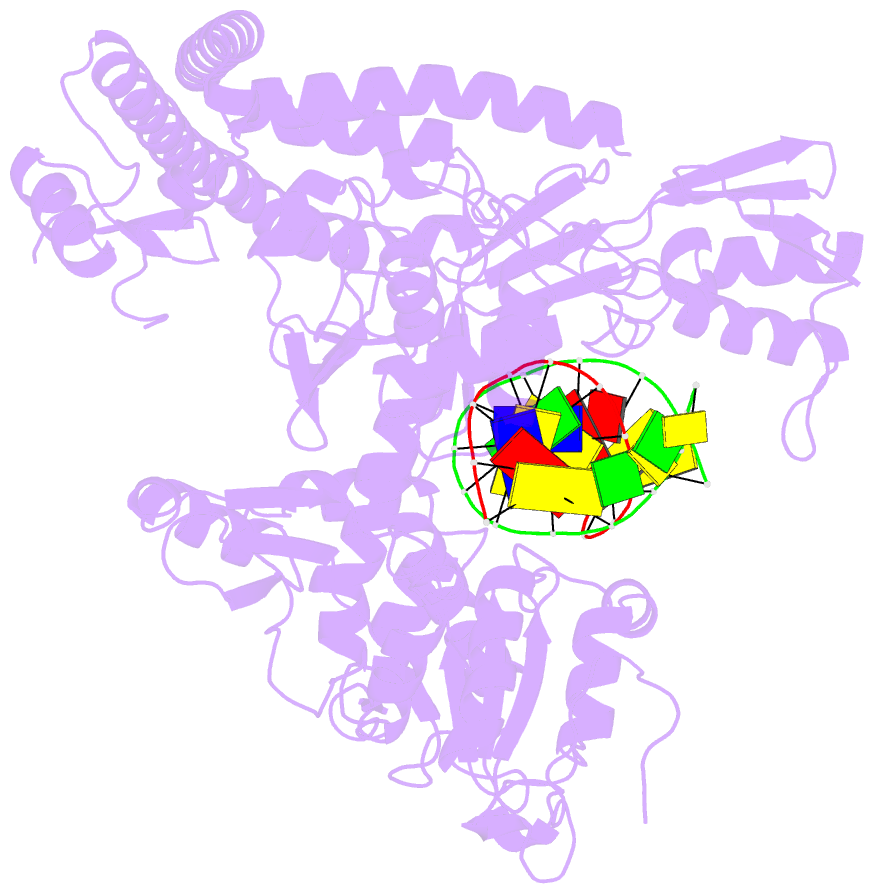
Summary information and primary citation
- PDB-id
- 3l4j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (2.48 Å)
- Summary
- Topoisomerase ii-DNA cleavage complex, apo
- Reference
- Schmidt BH, Burgin AB, Deweese JE, Osheroff N, Berger JM (2010): "A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases." Nature, 465, 641-644. doi: 10.1038/nature08974.
- Abstract
- Type II topoisomerases are required for the management of DNA tangles and supercoils, and are targets of clinical antibiotics and anti-cancer agents. These enzymes catalyse the ATP-dependent passage of one DNA duplex (the transport or T-segment) through a transient, double-stranded break in another (the gate or G-segment), navigating DNA through the protein using a set of dissociable internal interfaces, or 'gates'. For more than 20 years, it has been established that a pair of dimer-related tyrosines, together with divalent cations, catalyse G-segment cleavage. Recent efforts have proposed that strand scission relies on a 'two-metal mechanism', a ubiquitous biochemical strategy that supports vital cellular processes ranging from DNA synthesis to RNA self-splicing. Here we present the structure of the DNA-binding and cleavage core of Saccharomyces cerevisiae topoisomerase II covalently linked to DNA through its active-site tyrosine at 2.5A resolution, revealing for the first time the organization of a cleavage-competent type II topoisomerase configuration. Unexpectedly, metal-soaking experiments indicate that cleavage is catalysed by a novel variation of the classic two-metal approach. Comparative analyses extend this scheme to explain how distantly-related type IA topoisomerases cleave single-stranded DNA, unifying the cleavage mechanisms for these two essential enzyme families. The structure also highlights a hitherto undiscovered allosteric relay that actuates a molecular 'trapdoor' to prevent subunit dissociation during cleavage. This connection illustrates how an indispensable chromosome-disentangling machine auto-regulates DNA breakage to prevent the aberrant formation of mutagenic and cytotoxic genomic lesions.