Summary information and primary citation
- PDB-id
- 3laj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription regulator
- Method
- X-ray (2.306 Å)
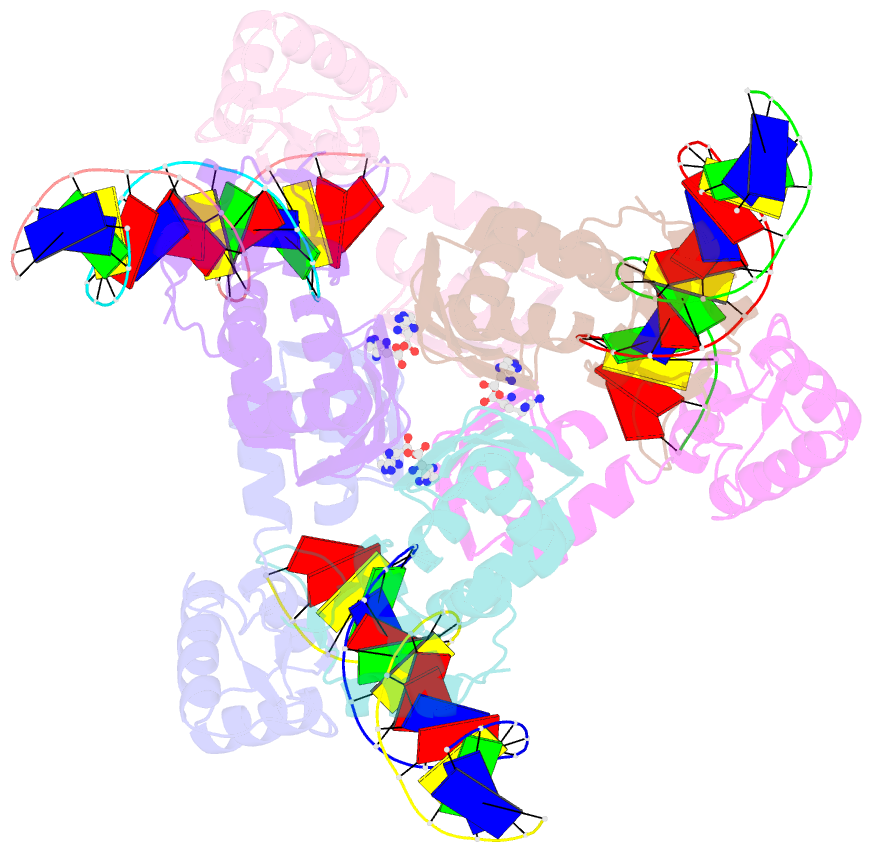
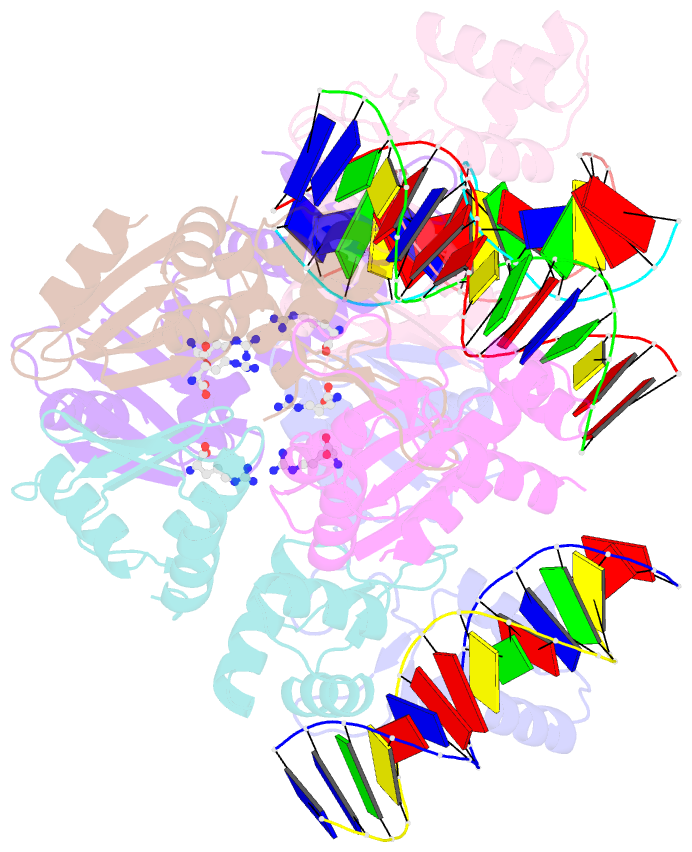
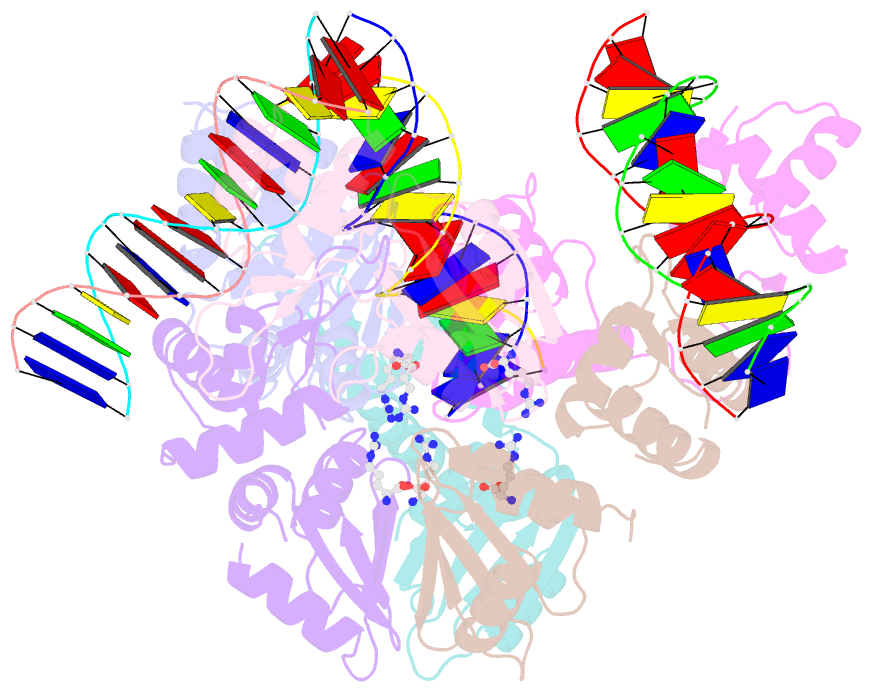
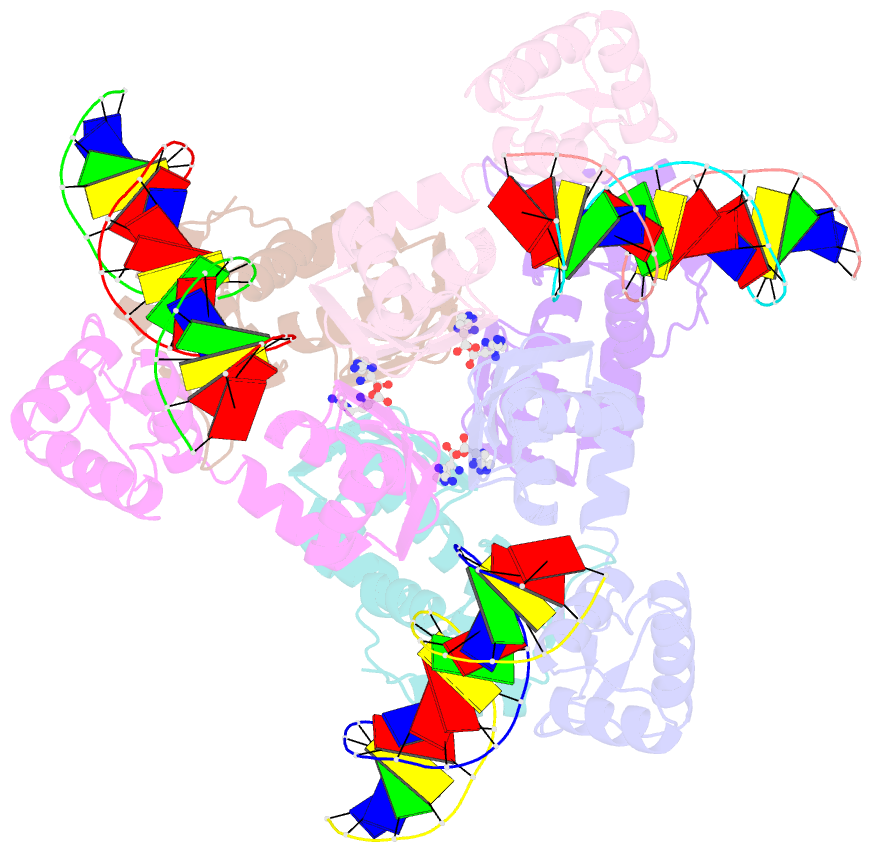
- Summary
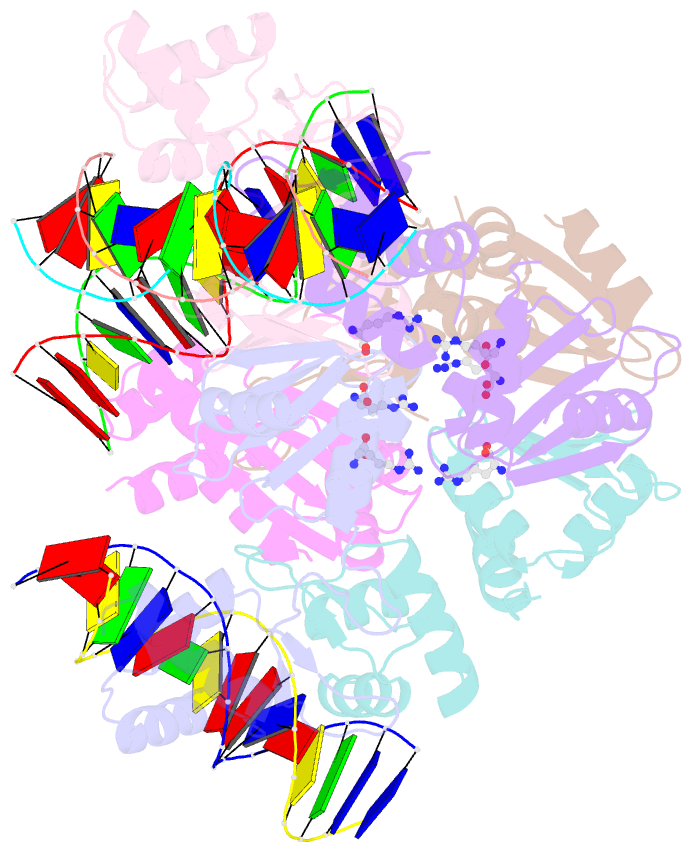
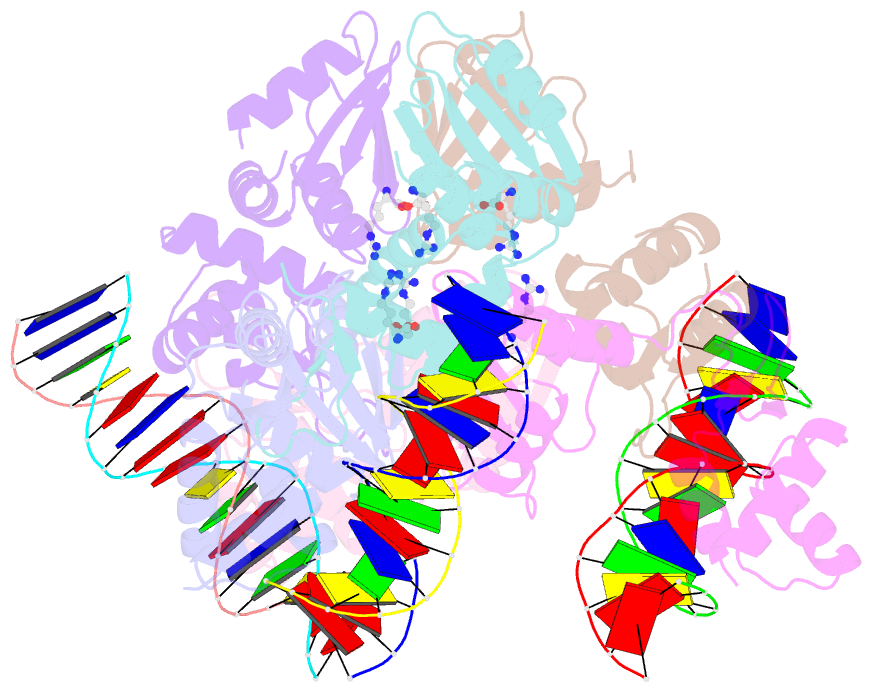
- The structure of the intermediate complex of the arginine repressor from mycobacterium tuberculosis bound to its DNA operator and l-arginine.
- Reference
- Cherney LT, Cherney MM, Garen CR, James MN (2010): "crystal structure of the intermediate complex of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator reveals detailed mechanism of arginine repression." J.Mol.Biol., 399, 240-254. doi: 10.1016/j.jmb.2010.03.065.
- Abstract
- The concentration of L-arginine in Mycobacterium tuberculosis (Mtb) and in many other bacteria is controlled by a transcriptional factor called the arginine repressor (ArgR). It regulates the transcription of the biosynthetic genes of the arginine operon by interacting with the approximately 16- to 20-bp ARG boxes in the promoter site of the operon. ArgRs in the arginine bound form are hexamers in which each protomer has two separately folded domains. The C-terminal domains form a hexameric core, whereas the N-terminal domains have the winged helix-turn-helix DNA-binding motif. Here, we present the crystal structure of the MtbArgR hexamer bound to three copies of the 16-bp DNA operator in the presence of trace amounts of L-arginine, determined to 2.15 A resolution. In contrast to our previously published structure of the ternary MtbArgR-DNA complex in the presence of 10 mM L-arginine, the DNA operators do not form a double ARG box in the structure reported here. The present structure not only retains the noncrystallographic 32 symmetry of the core (as in the earlier structure), but it also has the 3-fold axis for the whole complex. The core trimers are rotated relative to one another as in the other holo hexamers of MtbArgR, although the L-arginine ligands have only partial density and do not fully occupy the arginine-binding sites. Refinement of the occupancies and B-factors of ligands resulted in a value of approximately 4.4 arginine ligands per hexamer. This has allowed the dissociation constant of arginine from the arginine-binding site to be estimated. The present structure also has two protomer conformations, folded and extended. However, they are different from the conformations in the complex determined at an L-arginine concentration of 10 mM and do not form an interlocking arrangement. The new complex is less stable than the earlier described complex bound with nine arginine residues. Thus, the former can be considered as an intermediate in a pathway to the latter. On the basis of the structure of this intermediate complex, a more detailed mechanism of the arginine biosynthesis regulation in Mtb is proposed.