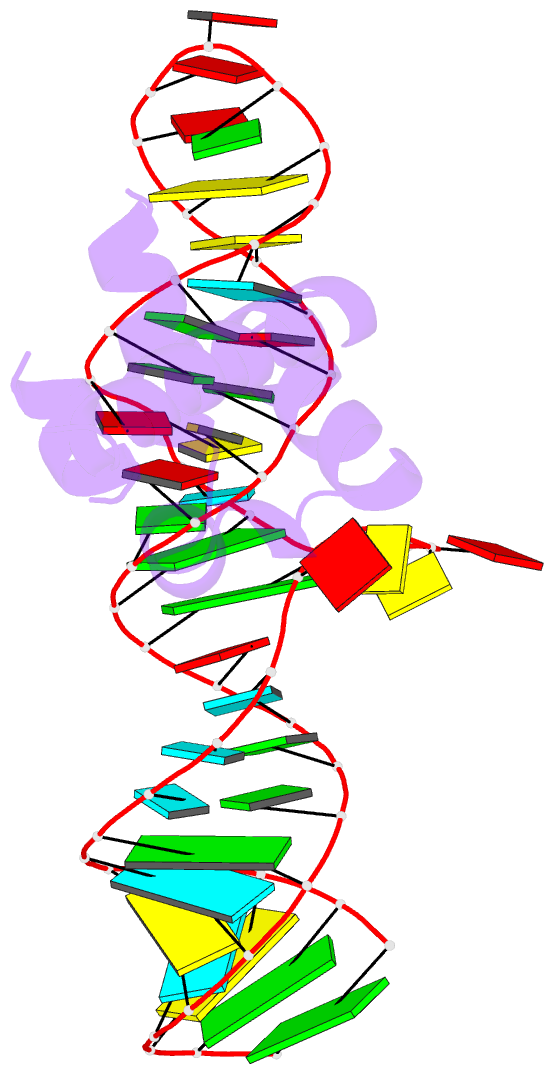
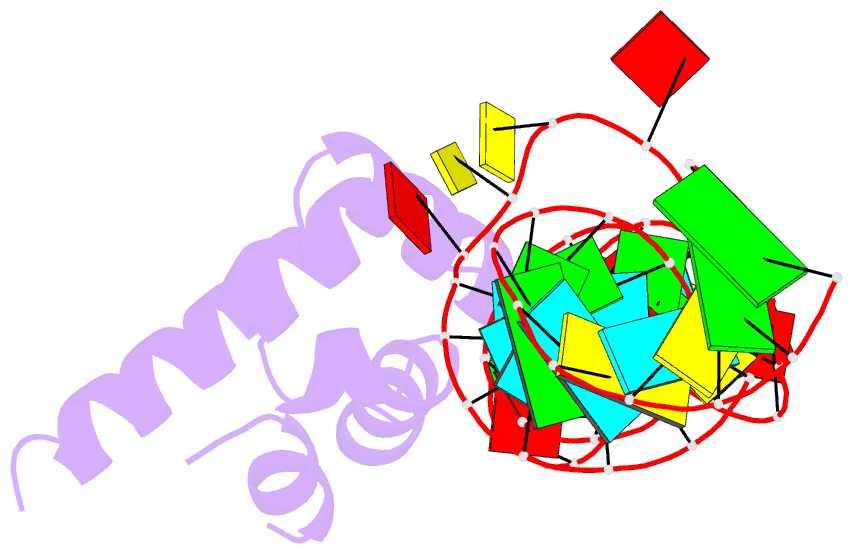
Summary information and primary citation
- PDB-id
- 3lqx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA-RNA binding protein
- Method
- X-ray (1.93 Å)
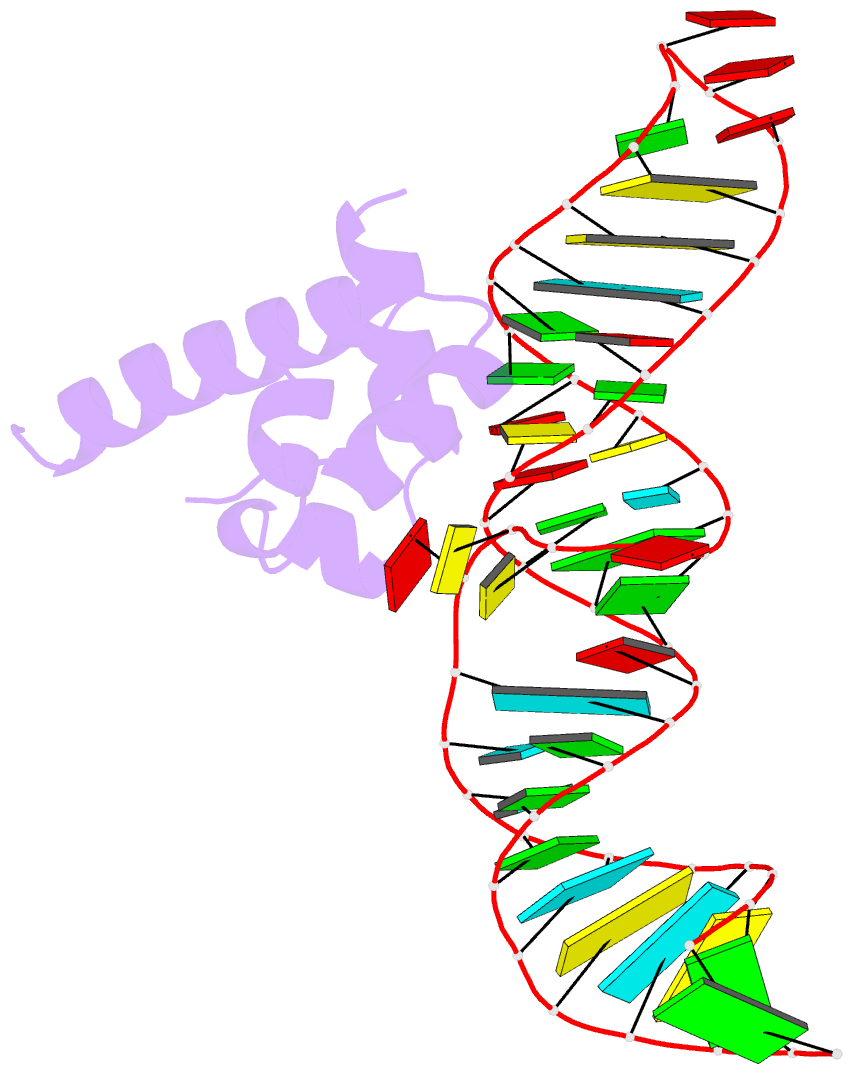
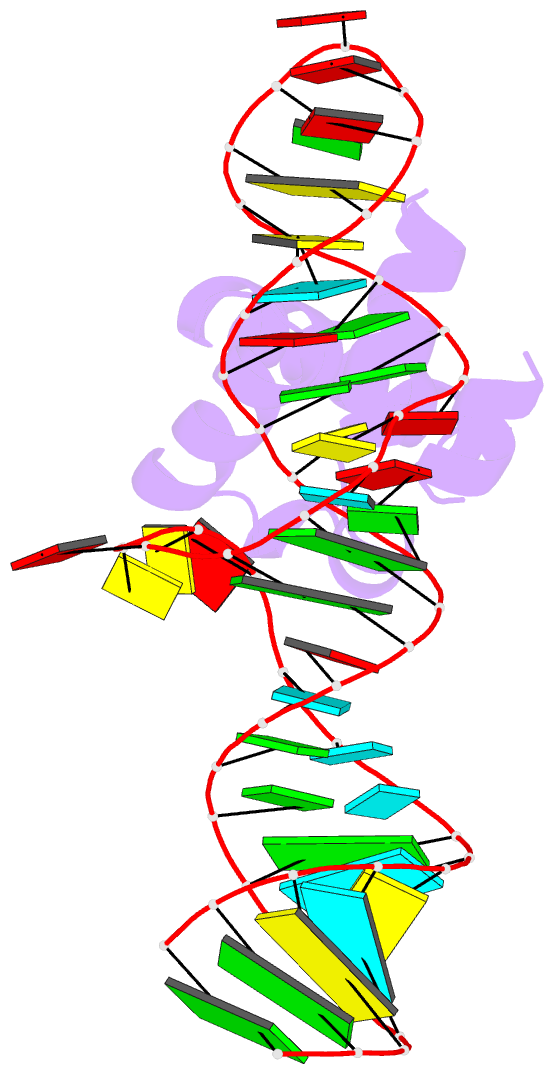
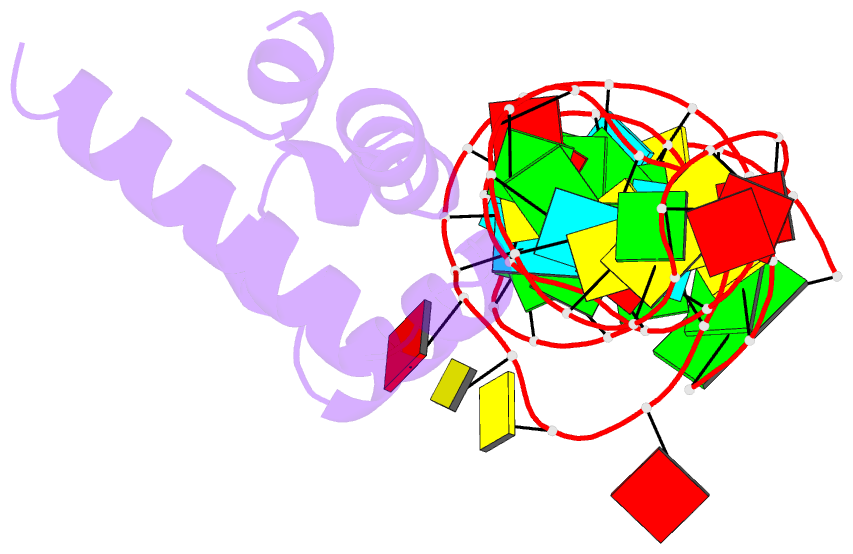
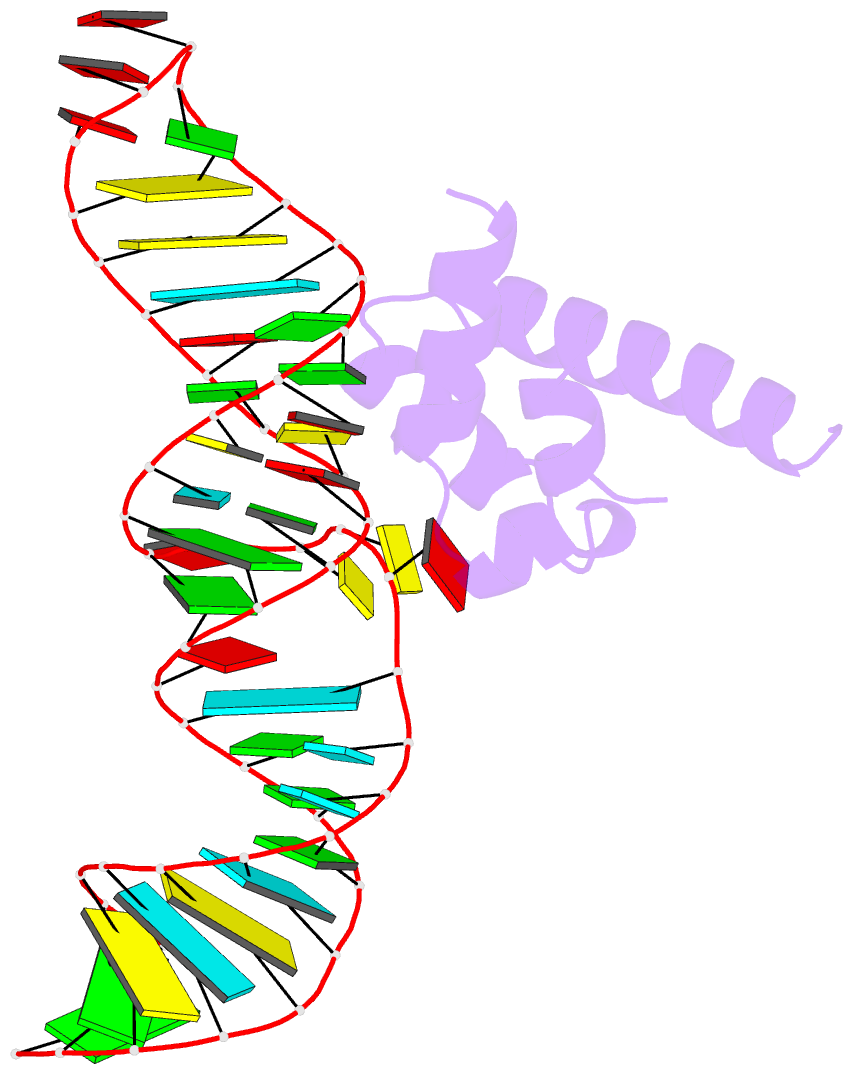
- Summary
- Srp ribonucleoprotein core complexed with cobalt hexammine
- Reference
- Batey RT, Doudna JA (2002): "Structural and Energetic Analysis of Metal Ions Essential to SRP Signal Recognition Domain Assembly." Biochemistry, 41, 11703-11710.
- Abstract
- The signal recognition particle (SRP) targets proteins to the endoplasmic reticulum in eukaryotes or to the inner membrane in prokaryotes by binding to hydrophobic signal sequences. Signal peptide recognition occurs within the highly conserved RNA-protein core of the SRP, underscoring the importance of this complex in SRP function. Structural analysis of the RNA and protein components of the prokaryotic SRP in the free and bound states revealed that the RNA undergoes a significant conformational change upon protein binding involving the uptake of several monovalent and divalent cations. To investigate the role of these metal ions in formation of the functional SRP complex, we used binding affinity assays and X-ray crystallography to analyze the specificity and energetic contributions of mono- and divalent metal ions bound in the RNA. Our results demonstrate that several metal ion binding sites important for RNA conformation can accommodate chemically distinct ions, often without affecting the structure of the complex. Thus, while these metal ions are highly ordered and essential for the formation and stability of the SRP complex, they behave like nonspecific metal ions.