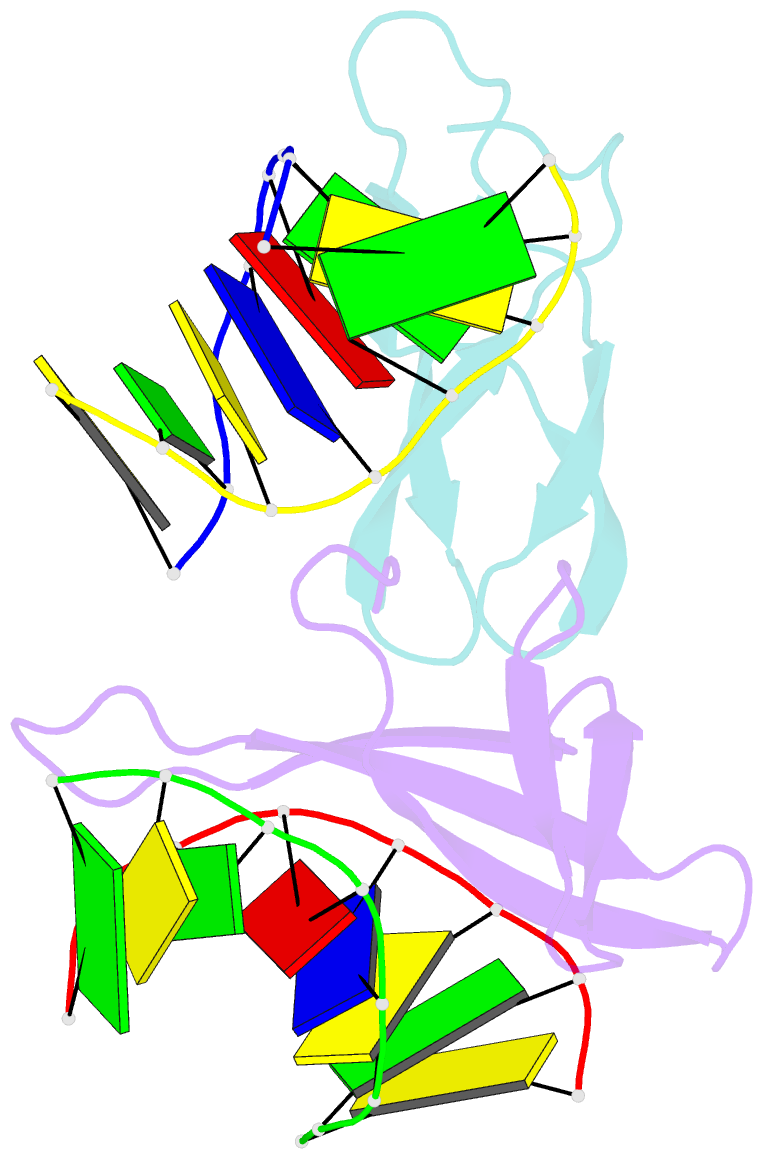
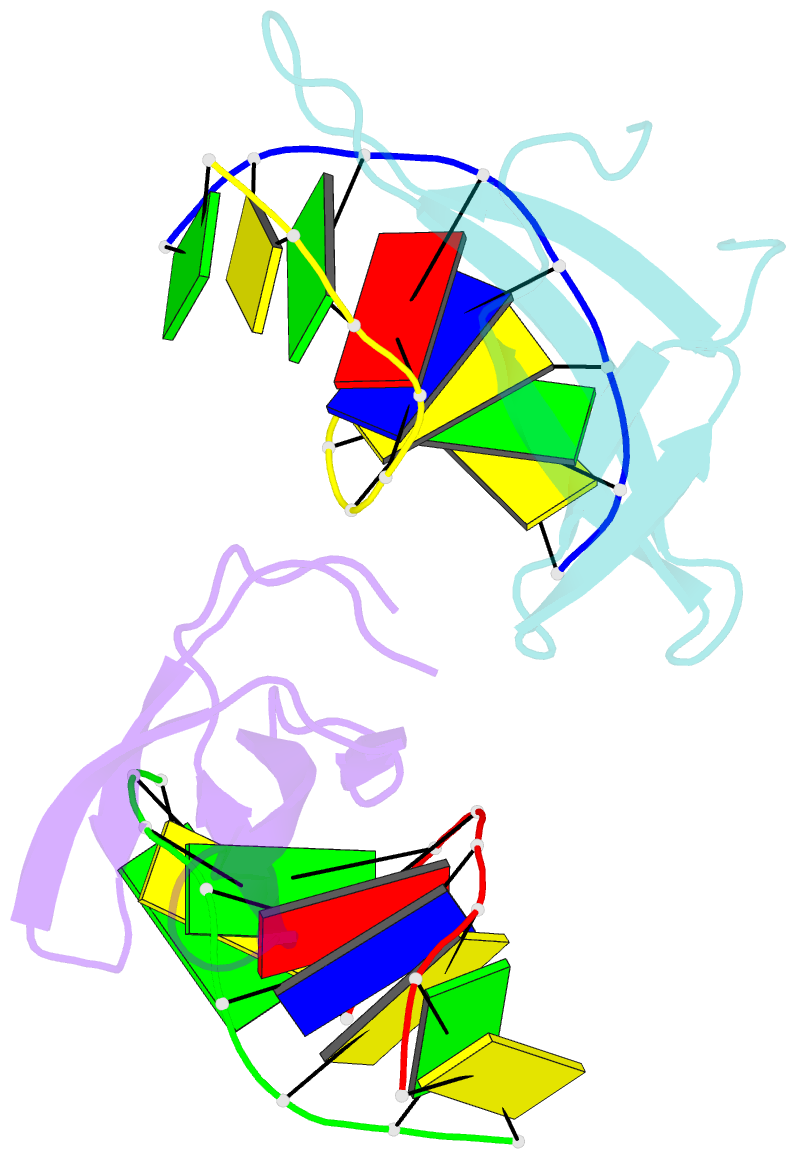
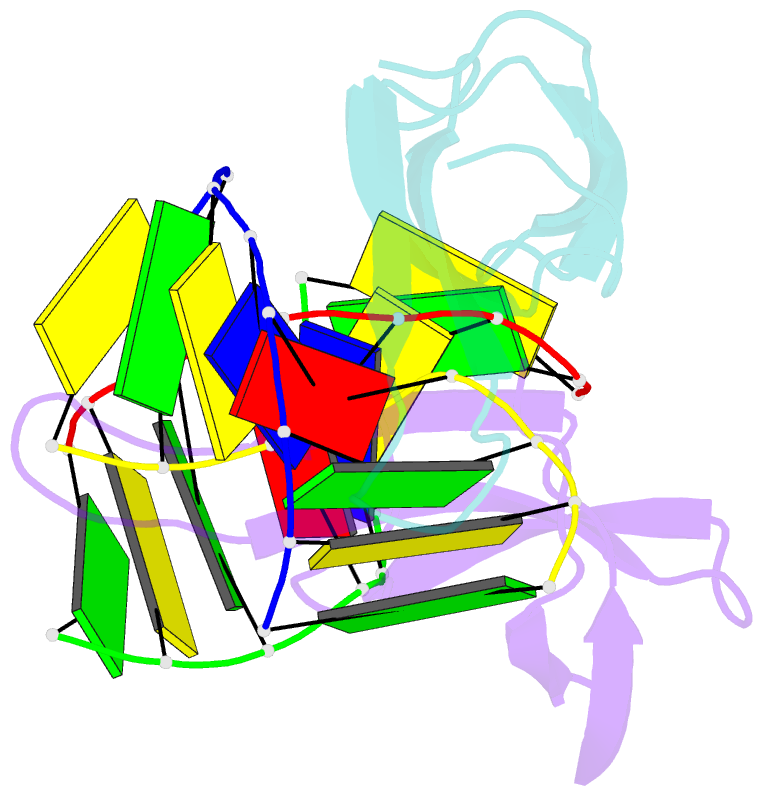
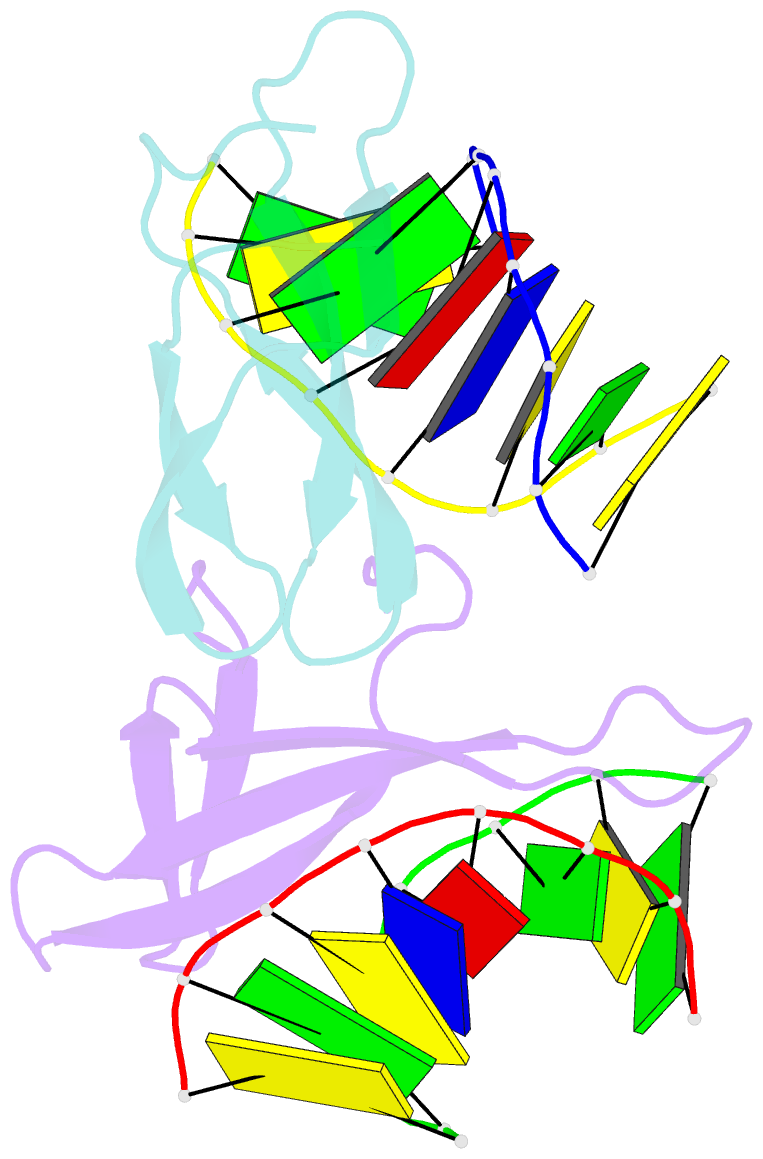
Summary information and primary citation
- PDB-id
- 3lwi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.3 Å)
- Summary
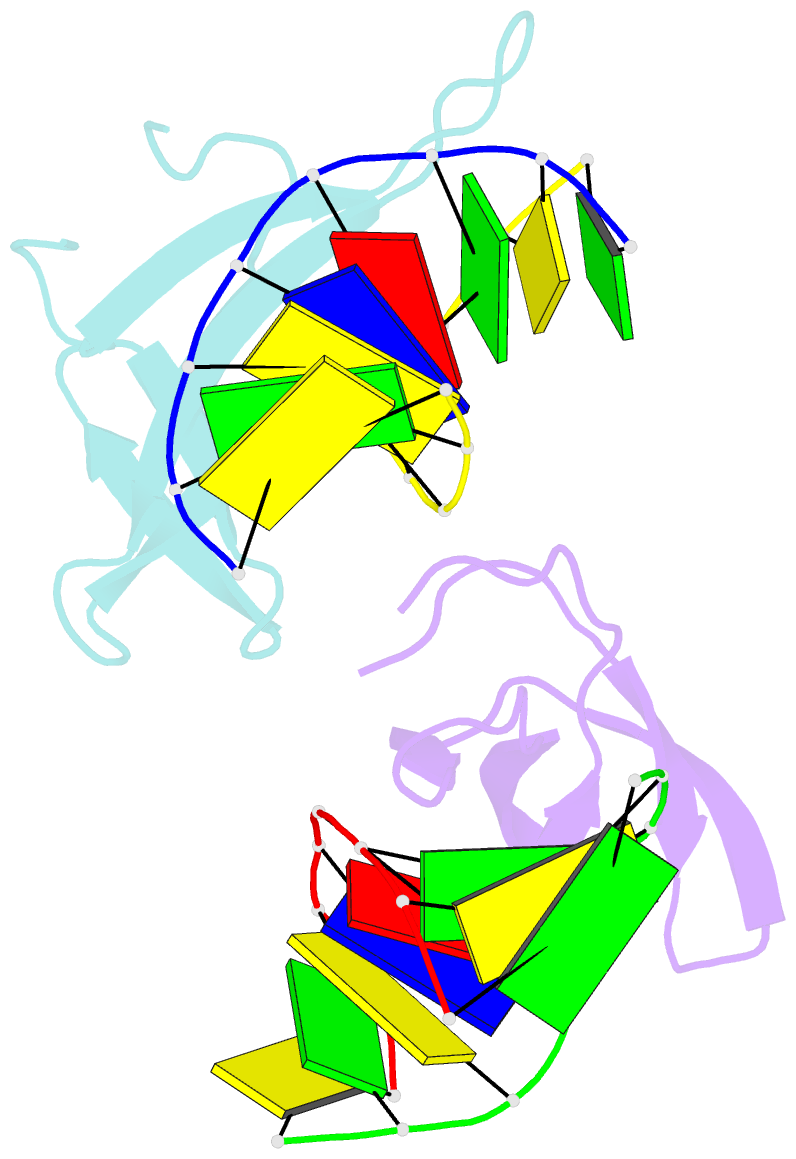
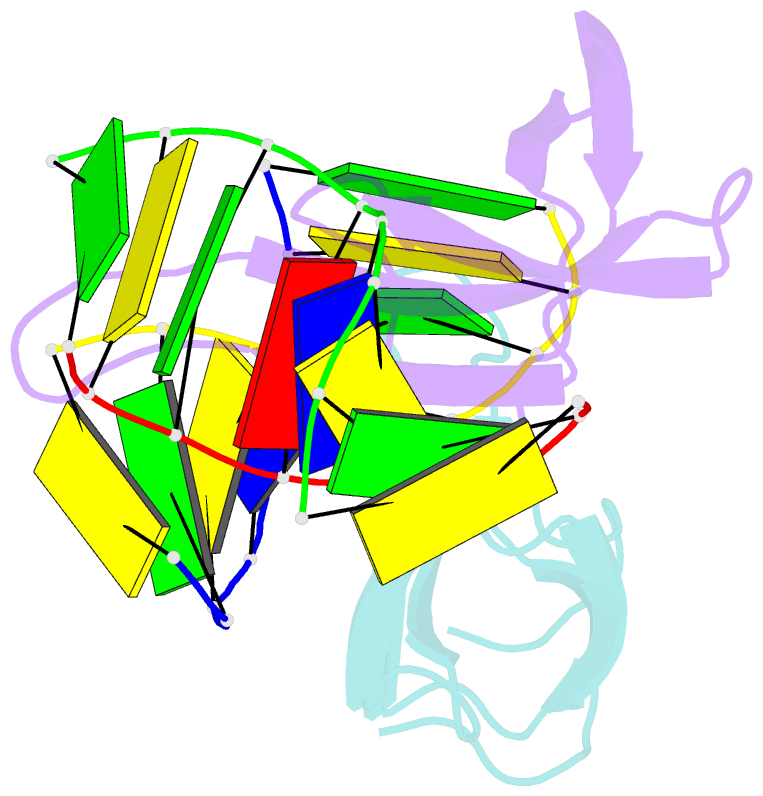
- Crystal structure of cren7-dsDNA complex
- Reference
- Zhang ZF, Gong Y, Guo L, Jiang T, Huang L (2010): "Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA." Mol.Microbiol., 76, 749-759. doi: 10.1111/j.1365-2958.2010.07136.x.
- Abstract
- Cren7, a newly found chromatin protein, is highly conserved in the Crenarchaeota. The protein shows higher affinity for double-stranded DNA than for single-stranded DNA, constrains negative DNA supercoils in vitro and is associated with genomic DNA in vivo. Here we report the crystal structures of the Cren7 protein from Sulfolobus solfataricus in complex with two DNA sequences. Cren7 binds in the minor groove of DNA and causes a single-step sharp kink in DNA (approximately 53 degrees) through the intercalation of the hydrophobic side chain of Leu28. Loop beta 3-beta 4 of Cren7 undergoes a significant conformational change upon binding of the protein to DNA, suggesting its critical role in the stabilization of the protein-DNA complex. The roles of DNA-contacting amino acid residues in stabilizing the Cren7-DNA interaction were examined by mutational analysis. Structural comparison of Cren7-DNA complexes with Sac7d-DNA complexes reveals significant differences between the two proteins in DNA binding surface, suggesting that Cren7 and Sul7d serve distinct functions in chromosomal organization.