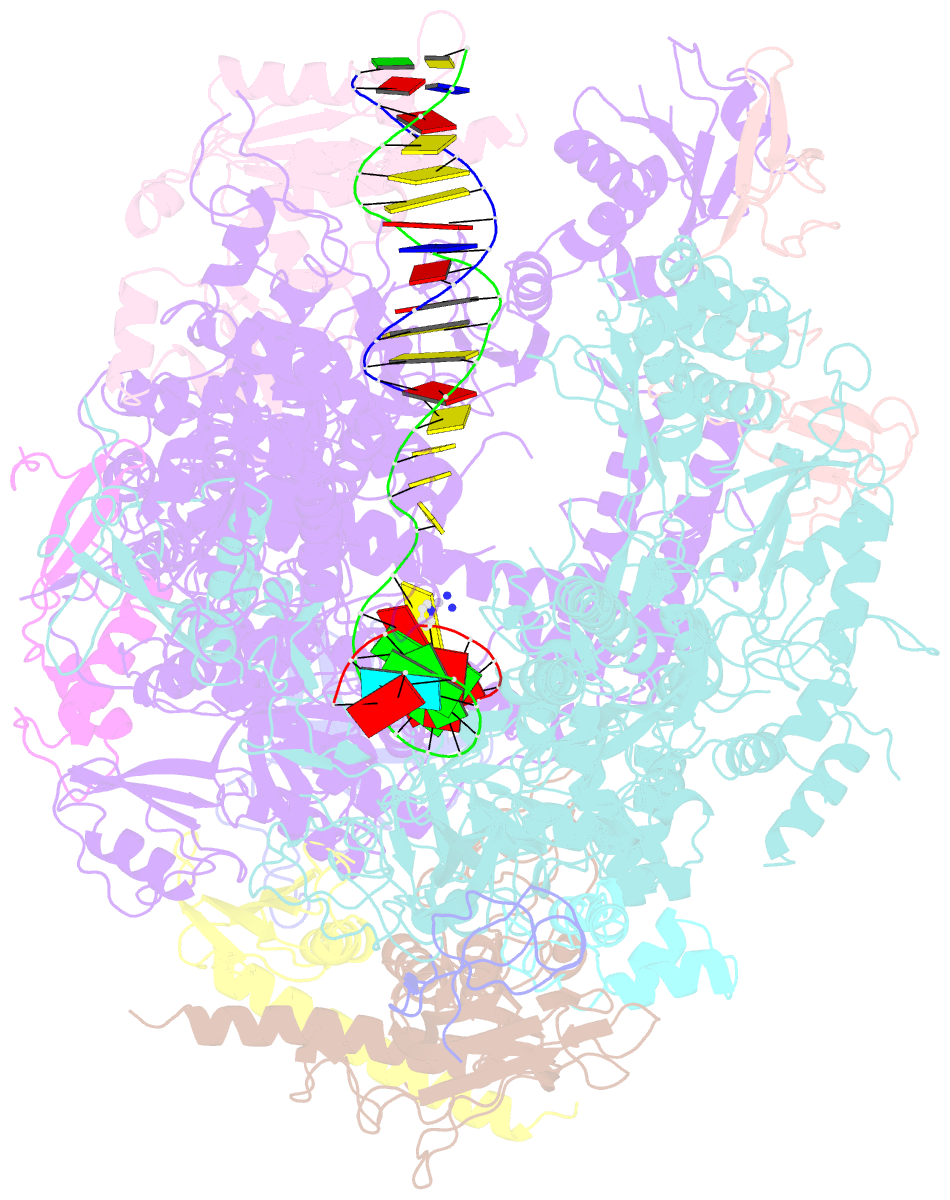
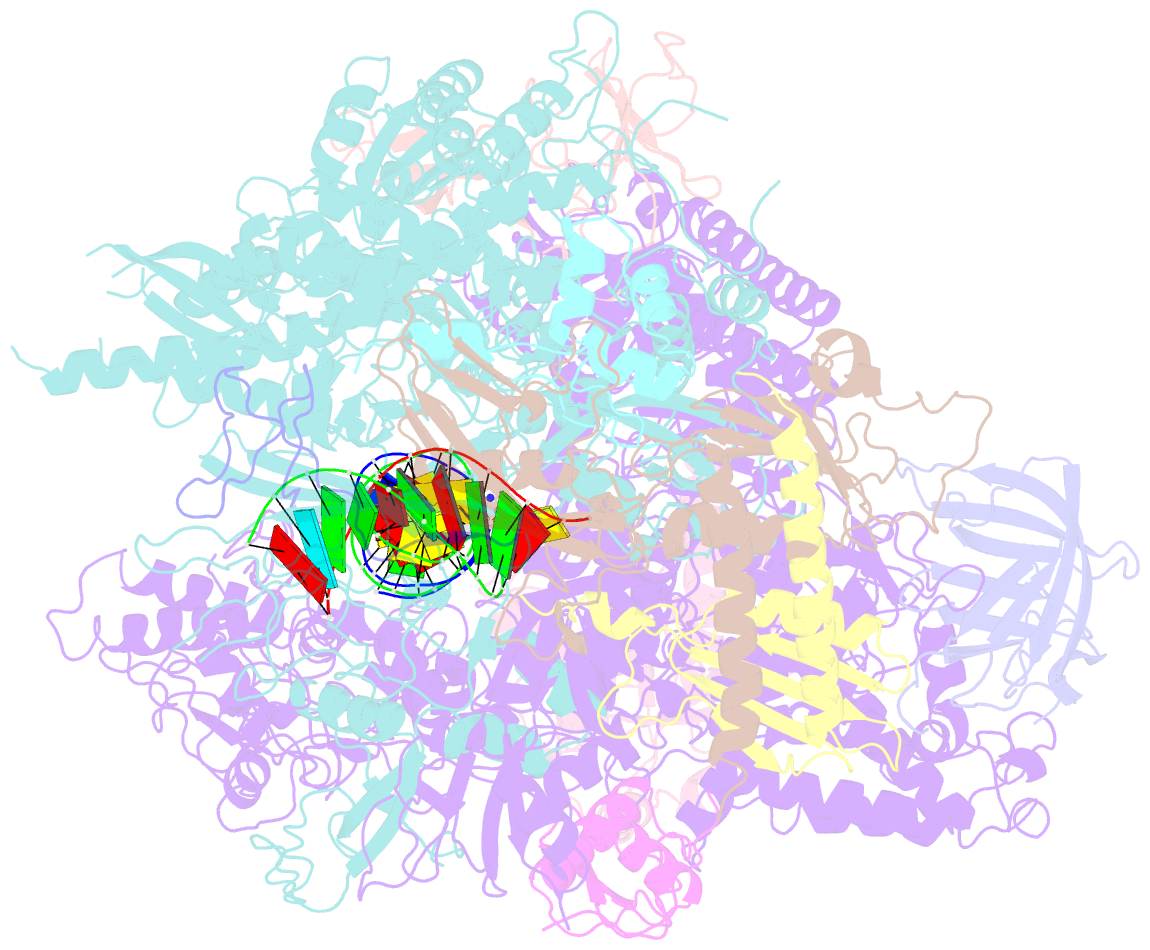
Summary information and primary citation
- PDB-id
- 3m3y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA hybrid
- Method
- X-ray (3.18 Å)
- Summary
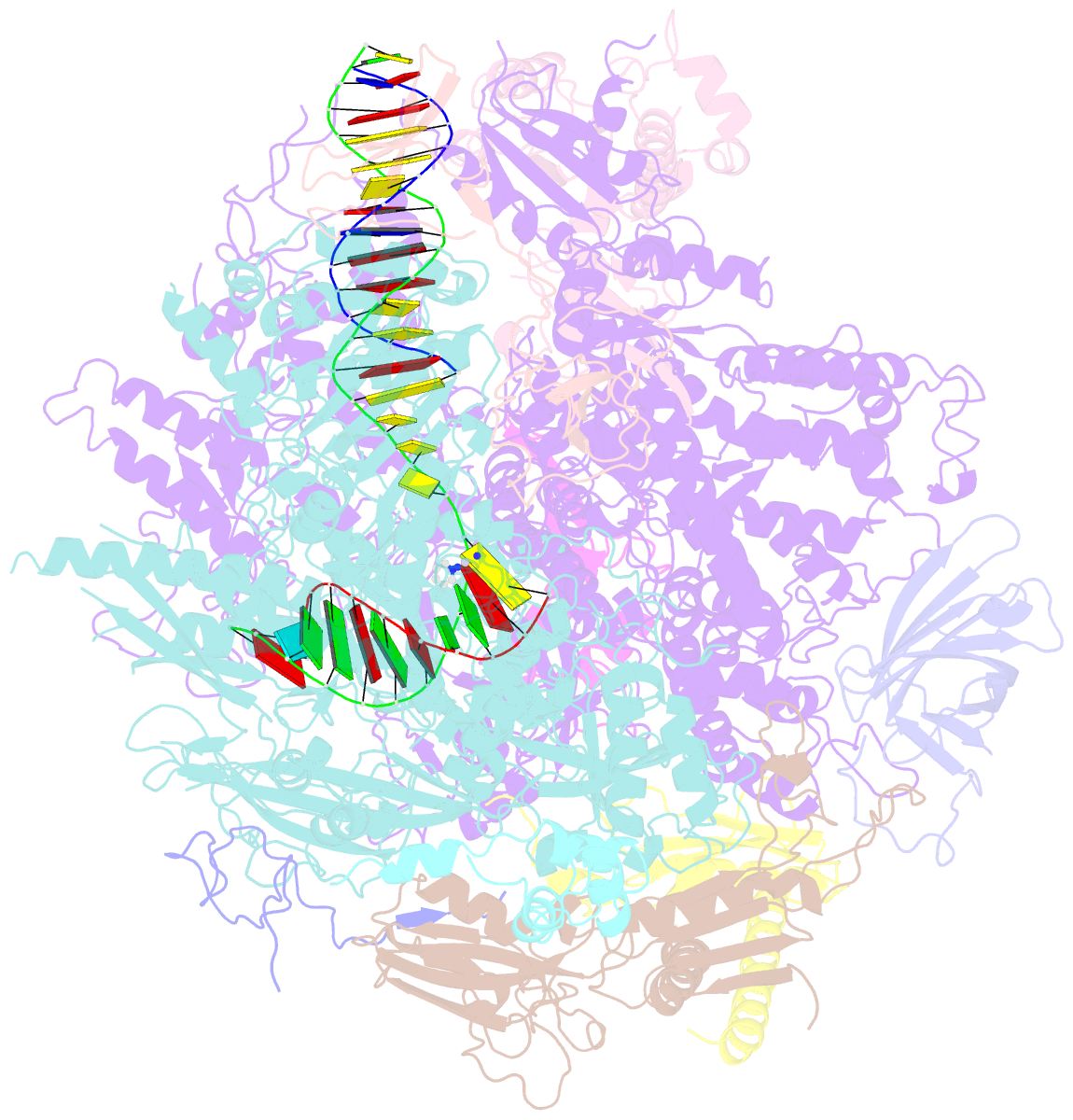
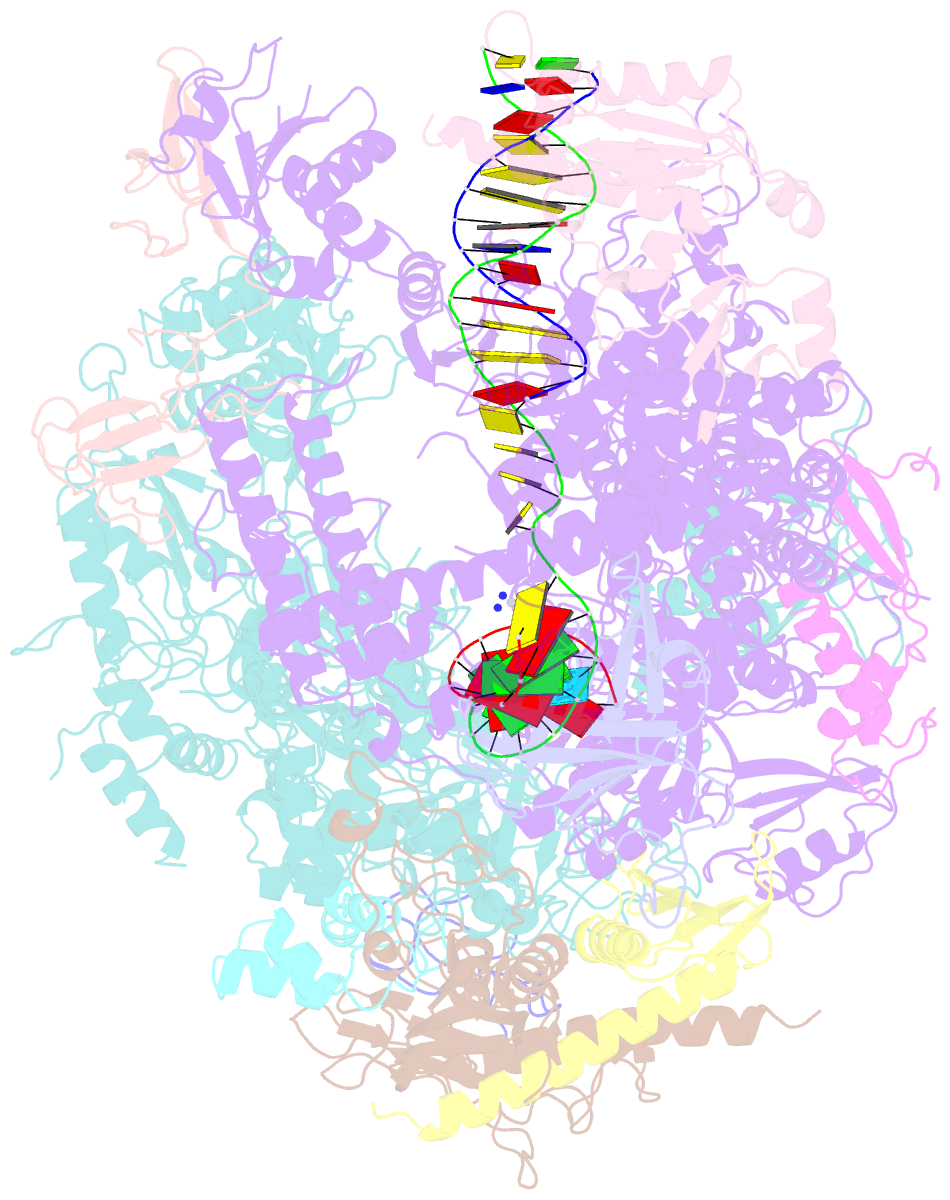
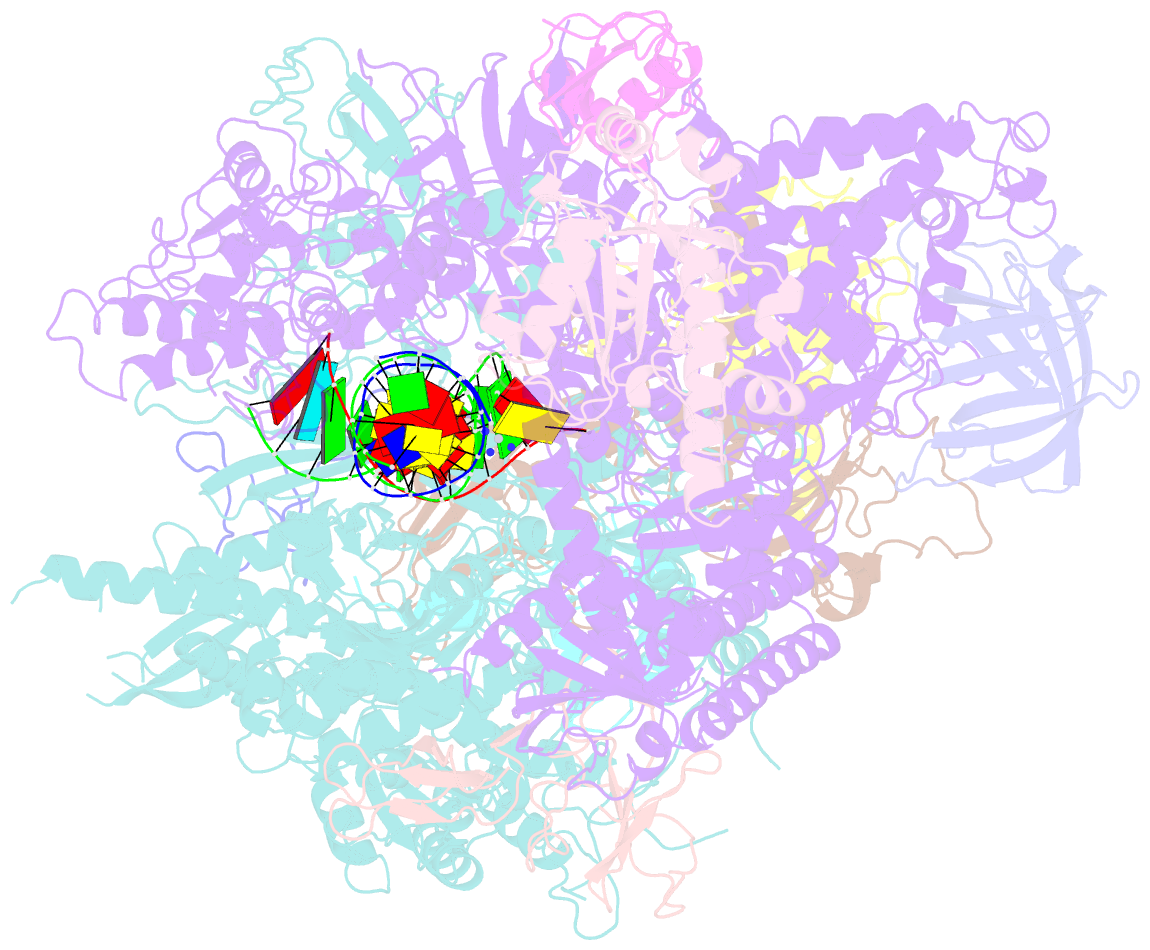
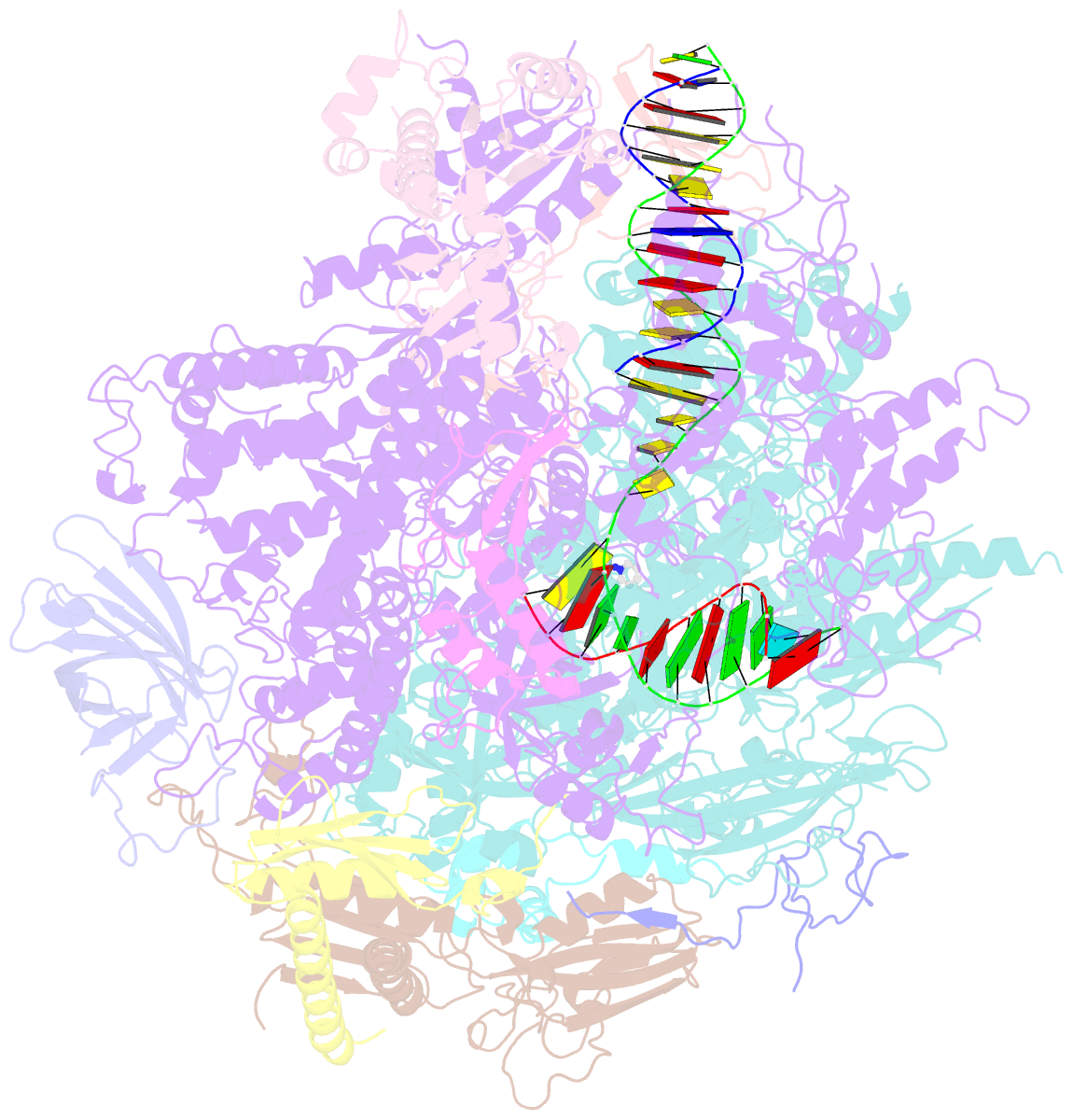
- RNA polymerase ii elongation complex c
- Reference
- Wang D, Zhu G, Huang X, Lippard SJ (2010): "X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct." Proc.Natl.Acad.Sci.USA, 107, 9584-9589. doi: 10.1073/pnas.1002565107.
- Abstract
- DNA is a major target of anticancer drugs. The resulting adducts interfere with key cellular processes, such as transcription, to trigger downstream events responsible for drug activity. cis-Diammine(pyridine)chloroplatinum(II), cDPCP or pyriplatin, is a monofunctional platinum(II) analogue of the widely used anticancer drug cisplatin having significant anticancer properties with a different spectrum of activity. Its novel structure-activity properties hold promise for overcoming drug resistance and improving the spectrum of treatable cancers over those responsive to cisplatin. However, the detailed molecular mechanism by which cells process DNA modified by pyriplatin and related monofunctional complexes is not at all understood. Here we report the structure of a transcribing RNA polymerase II (pol II) complex stalled at a site-specific monofunctional pyriplatin-DNA adduct in the active site. The results reveal a molecular mechanism of pol II transcription inhibition and drug action that is dramatically different from transcription inhibition by cisplatin and UV-induced 1,2-intrastrand cross-links. Our findings provide insight into structure-activity relationships that may apply to the entire family of monofunctional DNA-damaging agents and pave the way for rational improvement of monofunctional platinum anticancer drugs.