Summary information and primary citation
- PDB-id
- 3m4a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA
- Method
- X-ray (1.65 Å)
- Summary
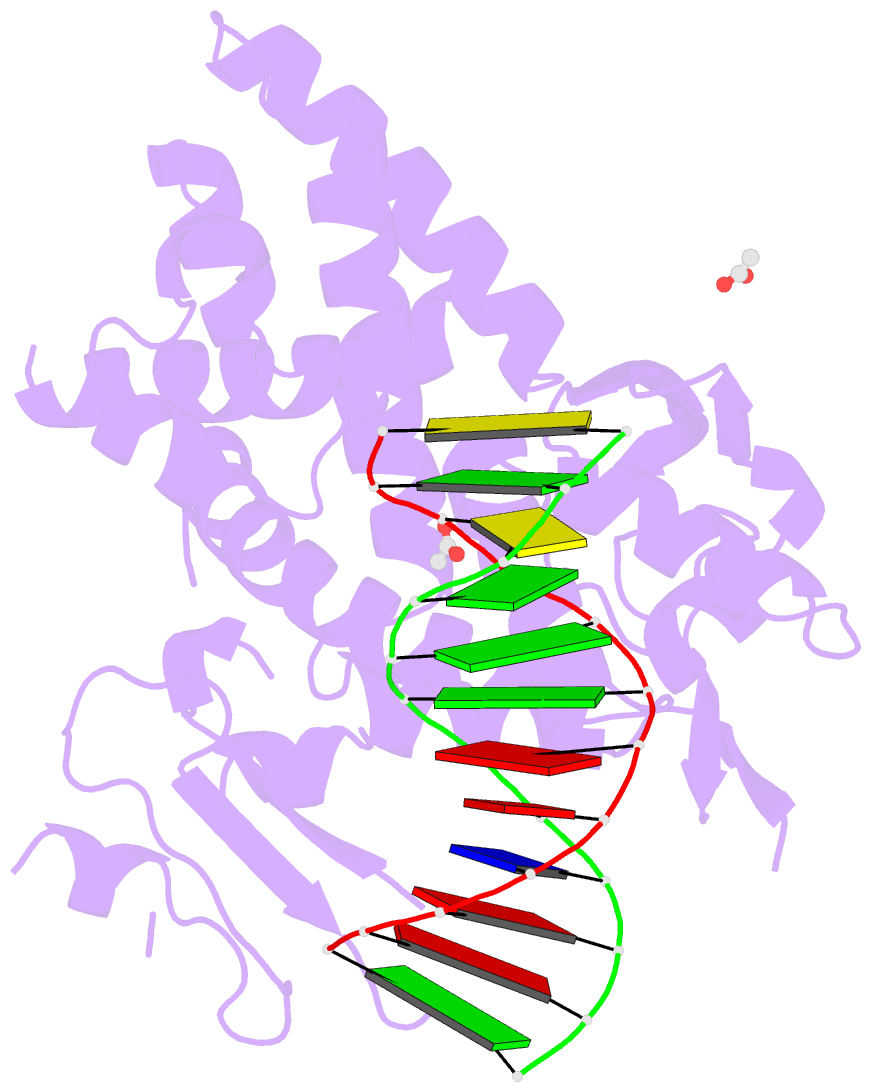
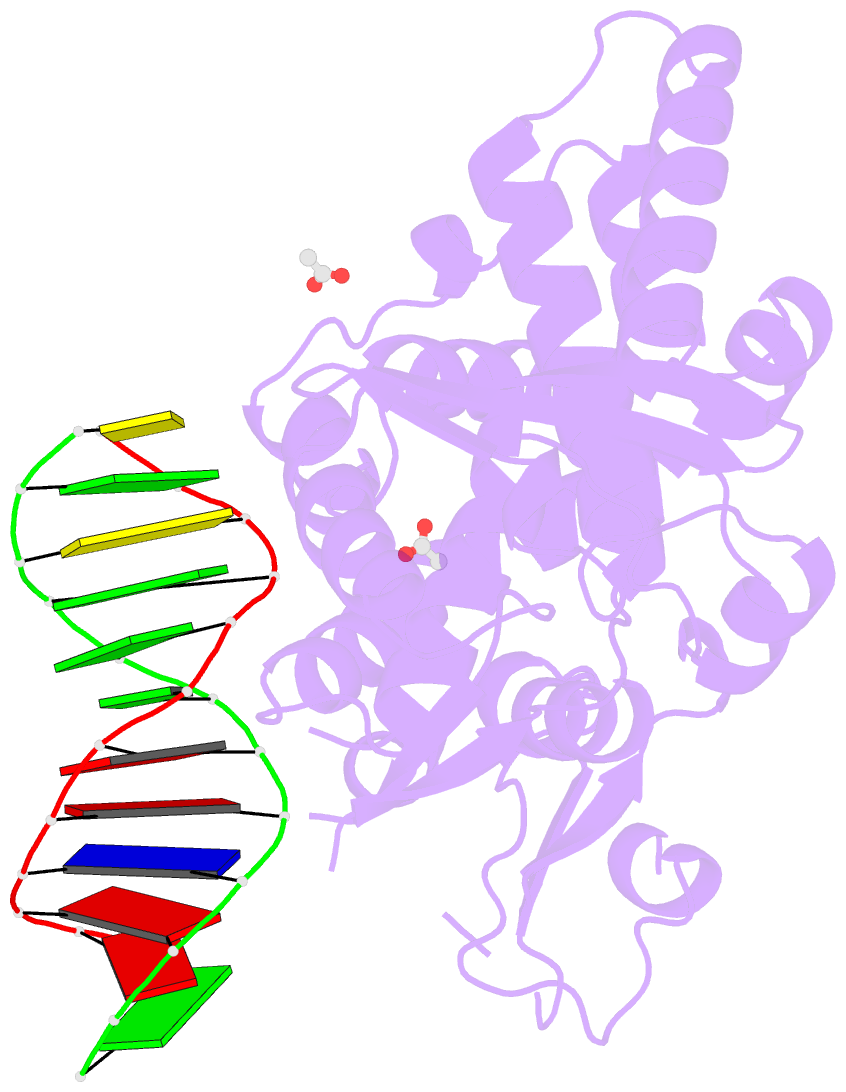
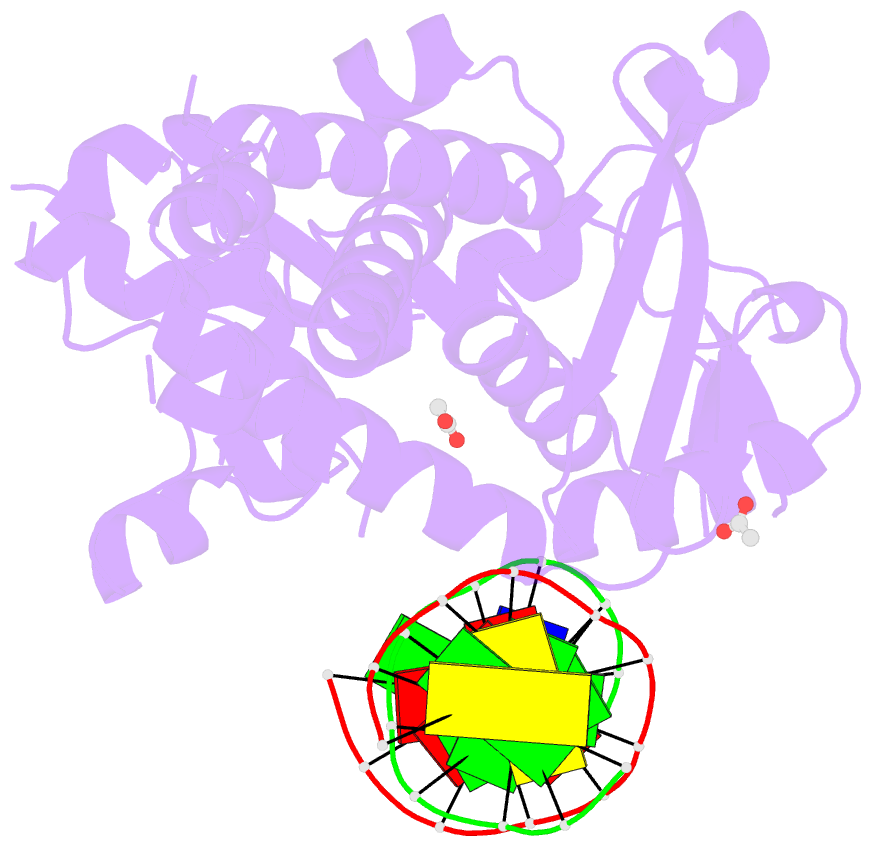
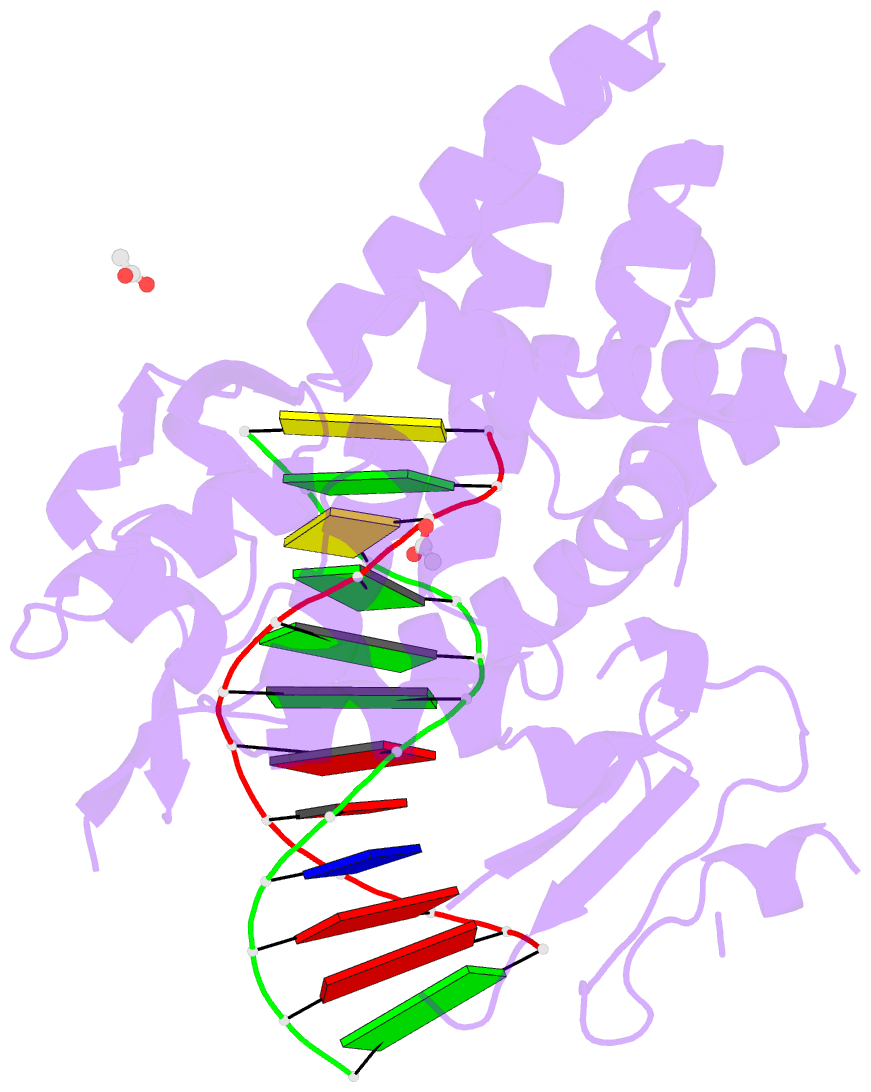
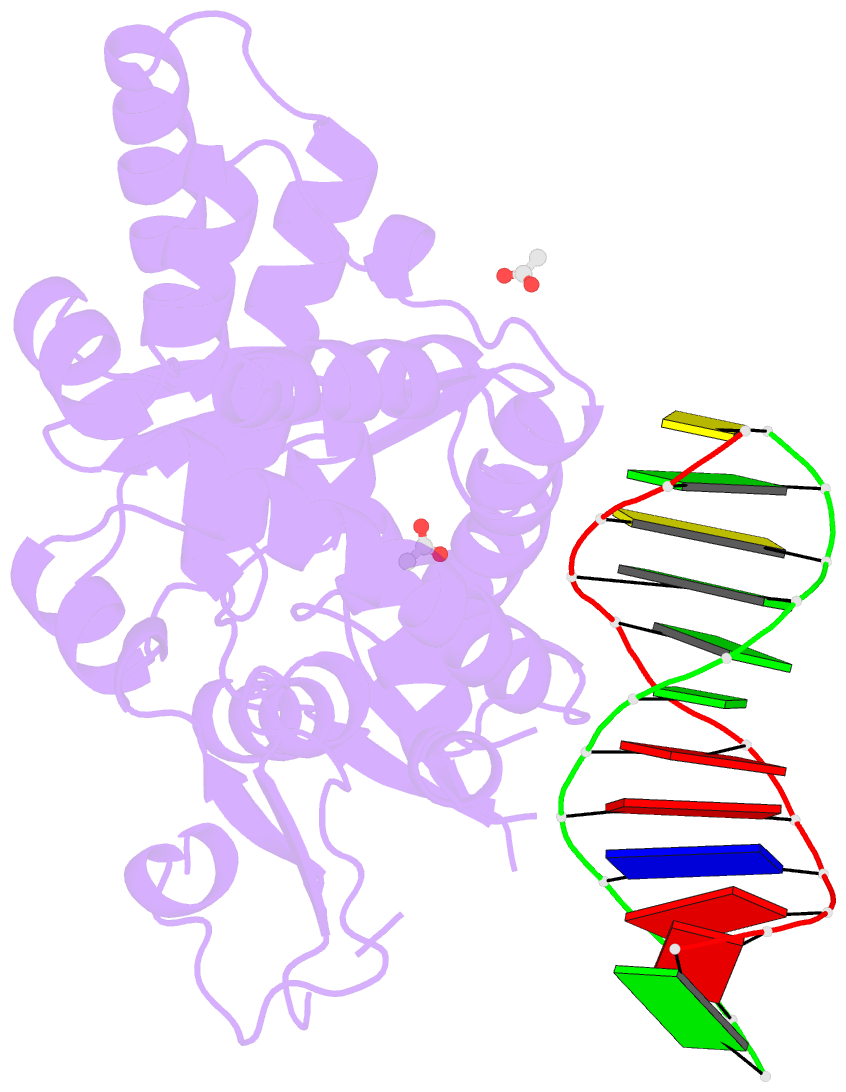
- Crystal structure of a bacterial topoisomerase ib in complex with DNA reveals a secondary DNA binding site
- Reference
- Patel A, Yakovleva L, Shuman S, Mondragon A (2010): "Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site." Structure, 18, 725-733. doi: 10.1016/j.str.2010.03.007.
- Abstract
- Type IB DNA topoisomerases (TopIB) are monomeric enzymes that relax supercoils by cleaving and resealing one strand of duplex DNA within a protein clamp that embraces a approximately 21 DNA segment. A longstanding conundrum concerns the capacity of TopIB enzymes to stabilize intramolecular duplex DNA crossovers and form protein-DNA synaptic filaments. Here we report a structure of Deinococcus radiodurans TopIB in complex with a 12 bp duplex DNA that demonstrates a secondary DNA binding site located on the surface of the C-terminal domain. It comprises a distinctive interface with one strand of the DNA duplex and is conserved in all TopIB enzymes. Modeling of a TopIB with both DNA sites suggests that the secondary site could account for DNA crossover binding, nucleation of DNA synapsis, and generation of a filamentous plectoneme. Mutations of the secondary site eliminate synaptic plectoneme formation without affecting DNA cleavage or supercoil relaxation.