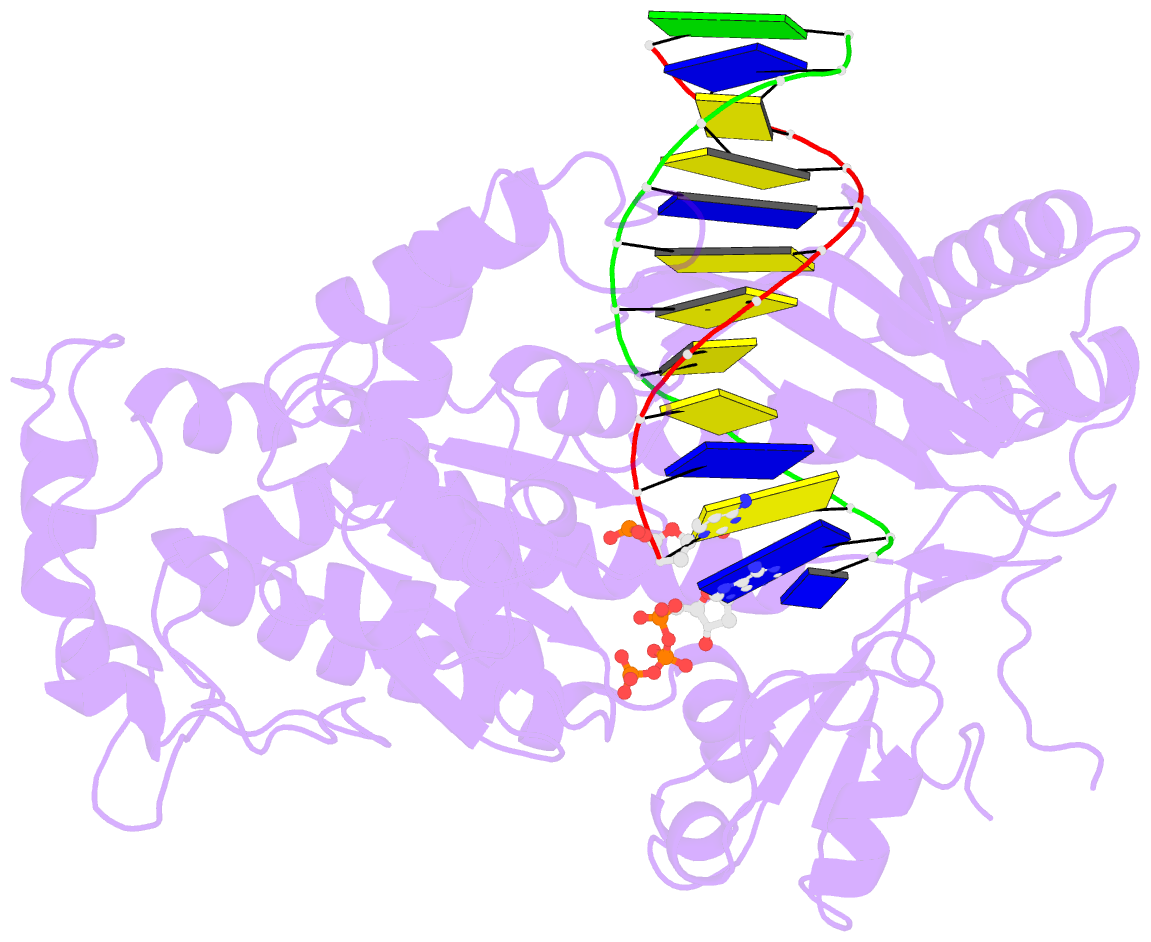
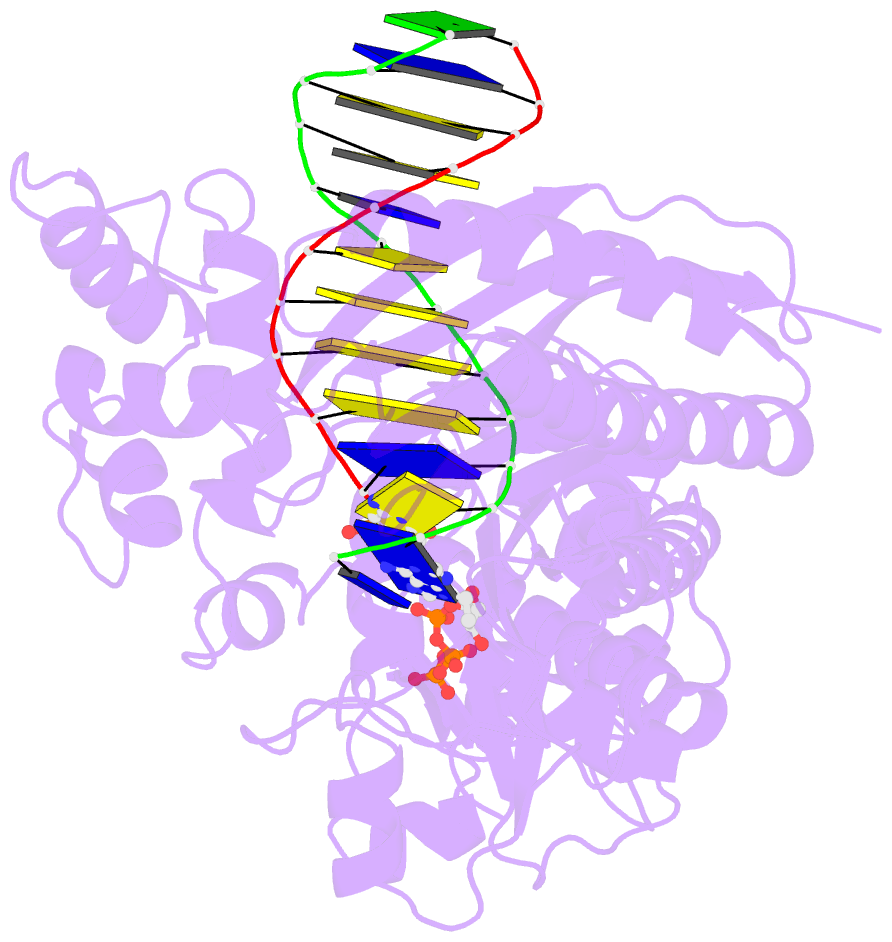
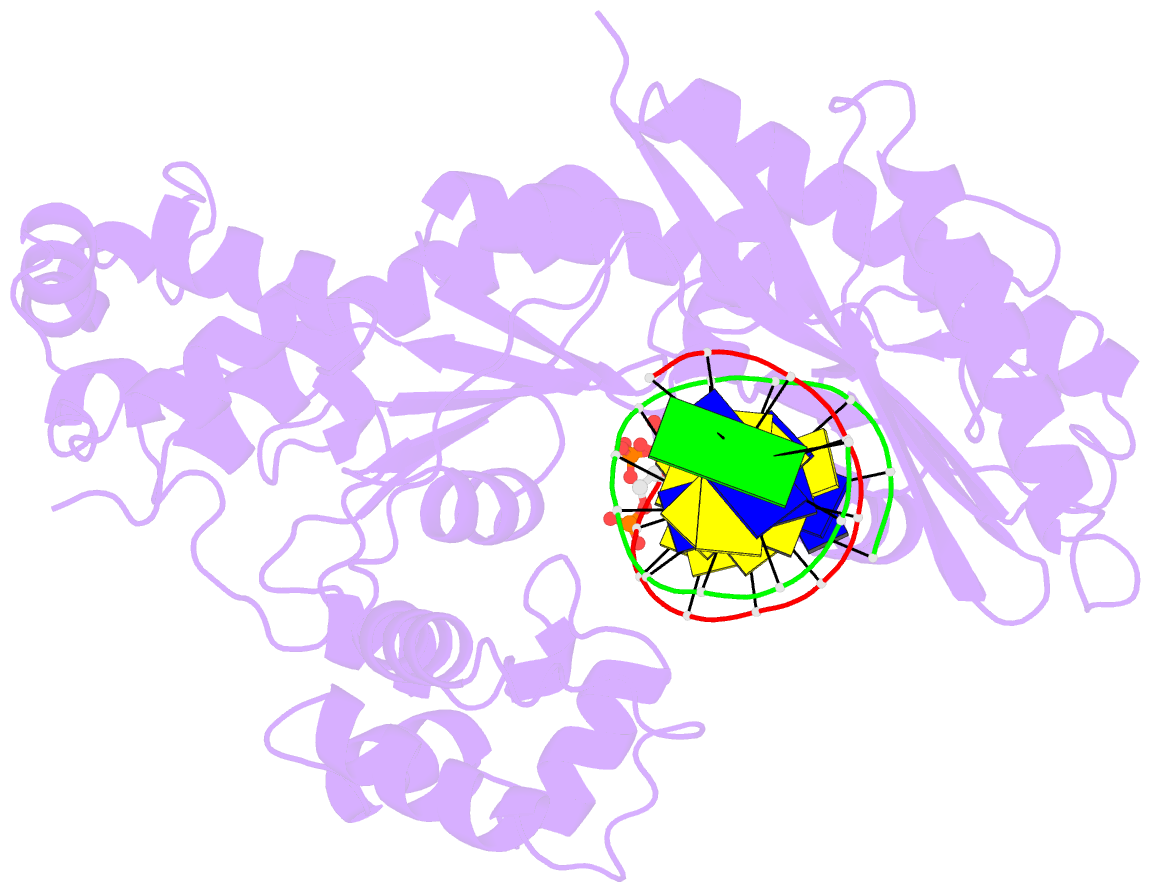
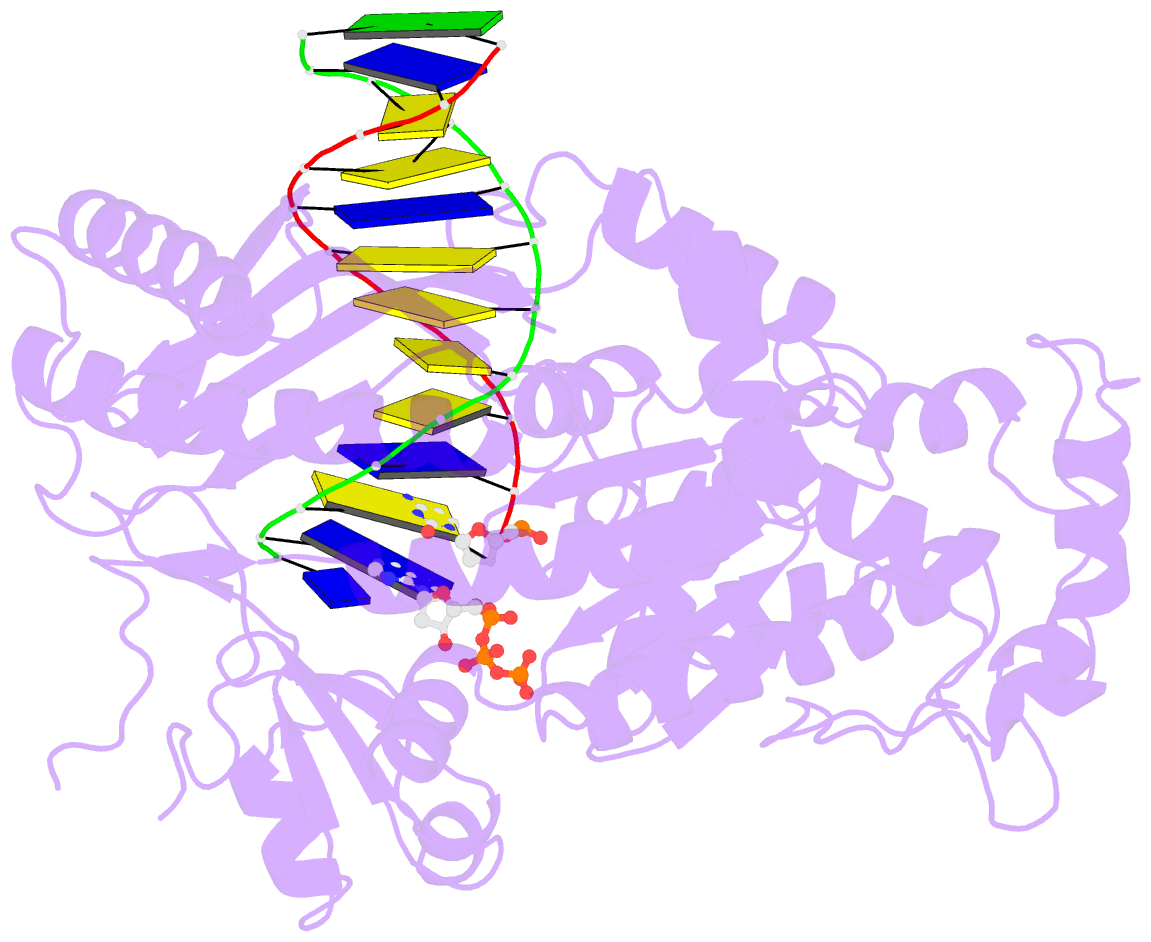
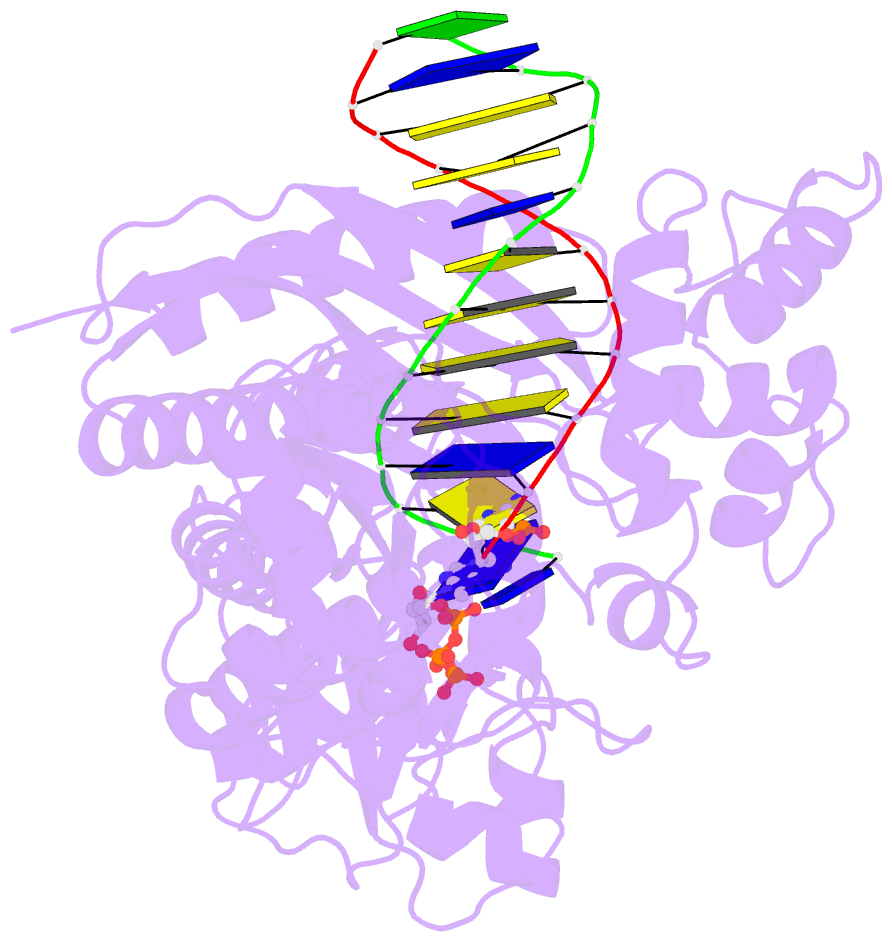
Summary information and primary citation
- PDB-id
- 3mfh; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
- DNA polymerase eta in complex with undamaged DNA
- Reference
- Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK (2010): "Structural basis for the suppression of skin cancers by DNA polymerase eta." Nature, 465, 1039-1043. doi: 10.1038/nature09104.
- Abstract
- DNA polymerase eta (Poleta) is unique among eukaryotic polymerases in its proficient ability for error-free replication through ultraviolet-induced cyclobutane pyrimidine dimers, and inactivation of Poleta (also known as POLH) in humans causes the variant form of xeroderma pigmentosum (XPV). We present the crystal structures of Saccharomyces cerevisiae Poleta (also known as RAD30) in ternary complex with a cis-syn thymine-thymine (T-T) dimer and with undamaged DNA. The structures reveal that the ability of Poleta to replicate efficiently through the ultraviolet-induced lesion derives from a simple and yet elegant mechanism, wherein the two Ts of the T-T dimer are accommodated in an active site cleft that is much more open than in other polymerases. We also show by structural, biochemical and genetic analysis that the two Ts are maintained in a stable configuration in the active site via interactions with Gln 55, Arg 73 and Met 74. Together, these features define the basis for Poleta's action on ultraviolet-damaged DNA that is crucial in suppressing the mutagenic and carcinogenic consequences of sun exposure, thereby reducing the incidence of skin cancers in humans.