Summary information and primary citation
- PDB-id
- 3mfk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.0 Å)
- Summary
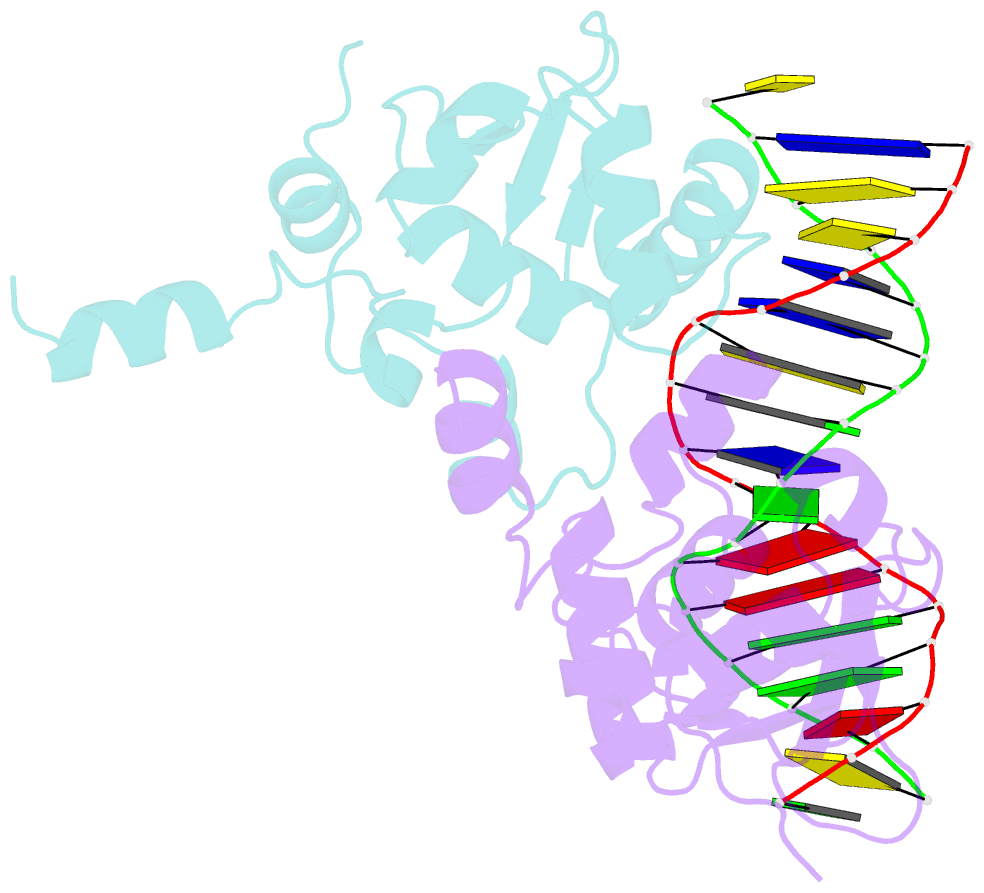
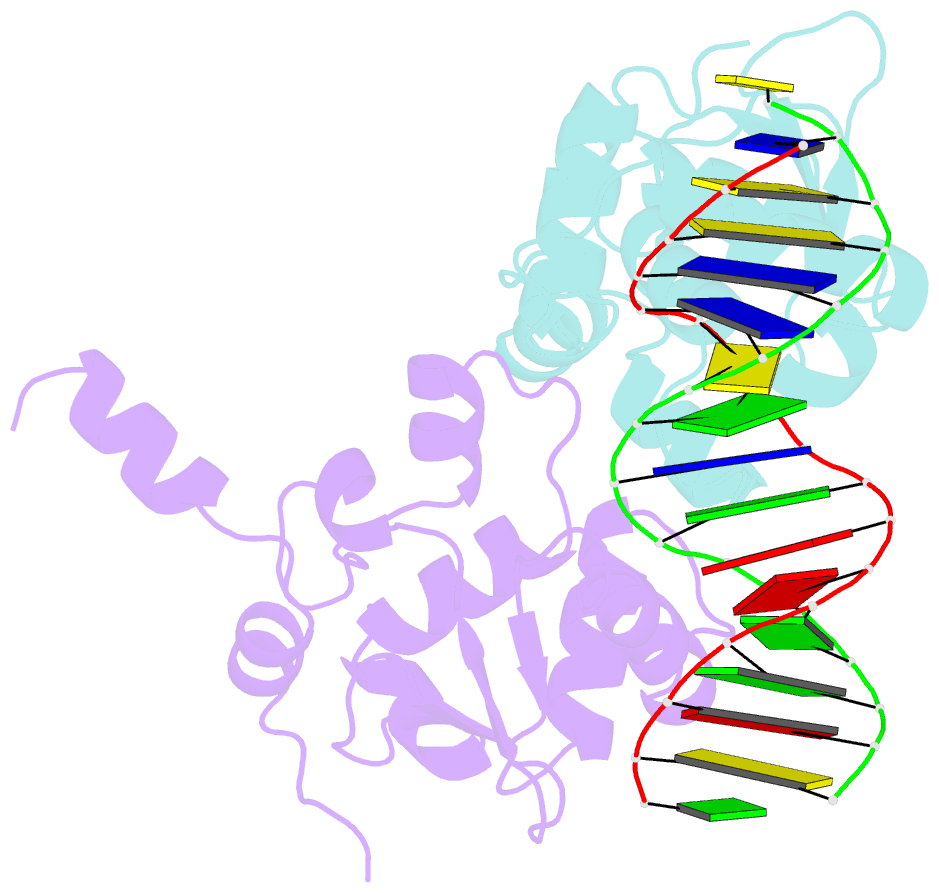
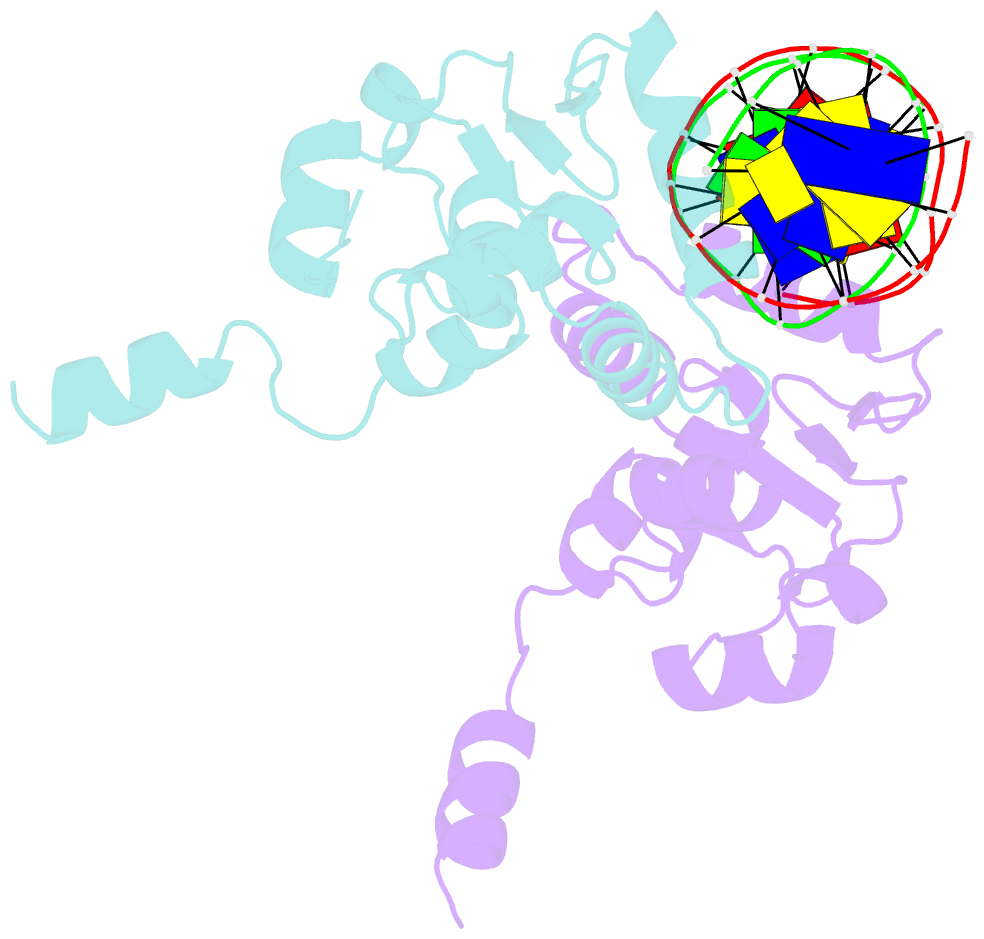
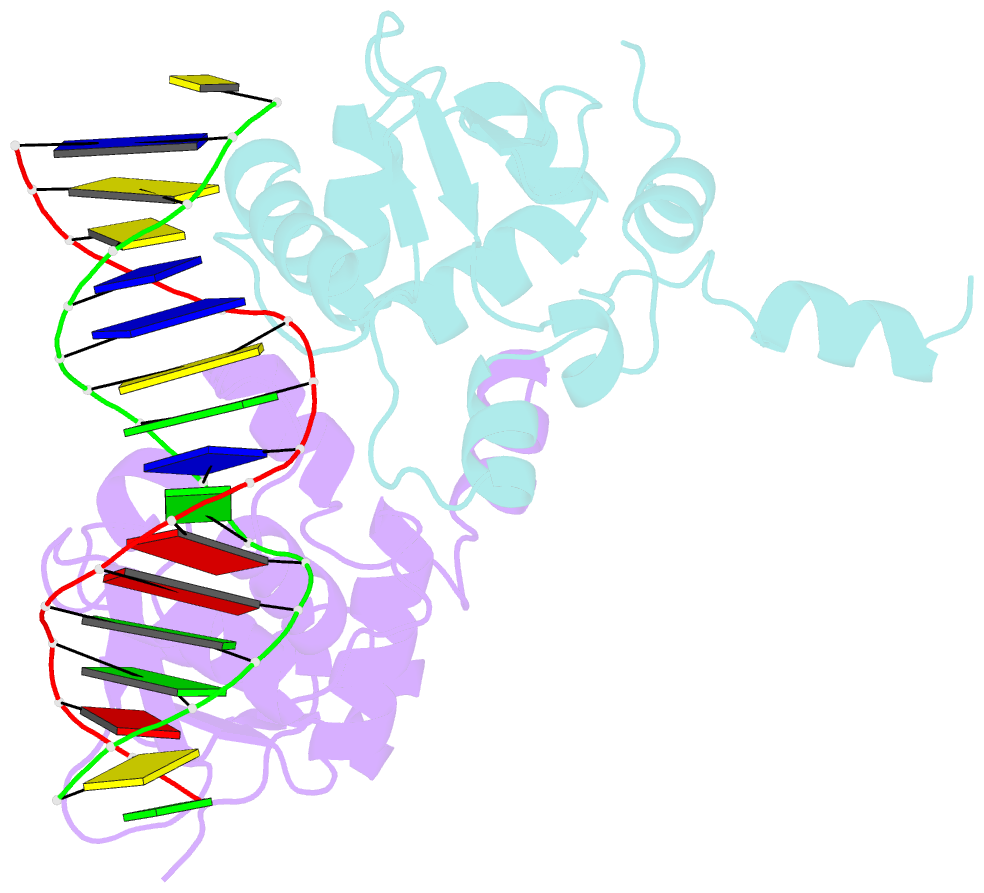
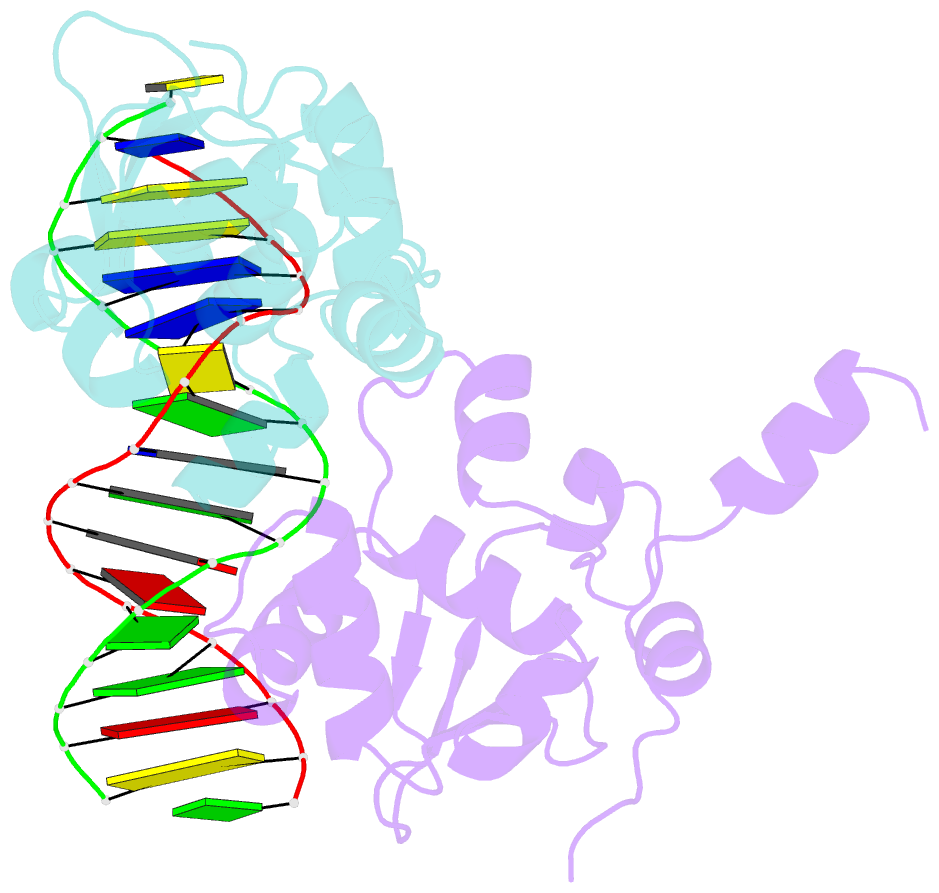
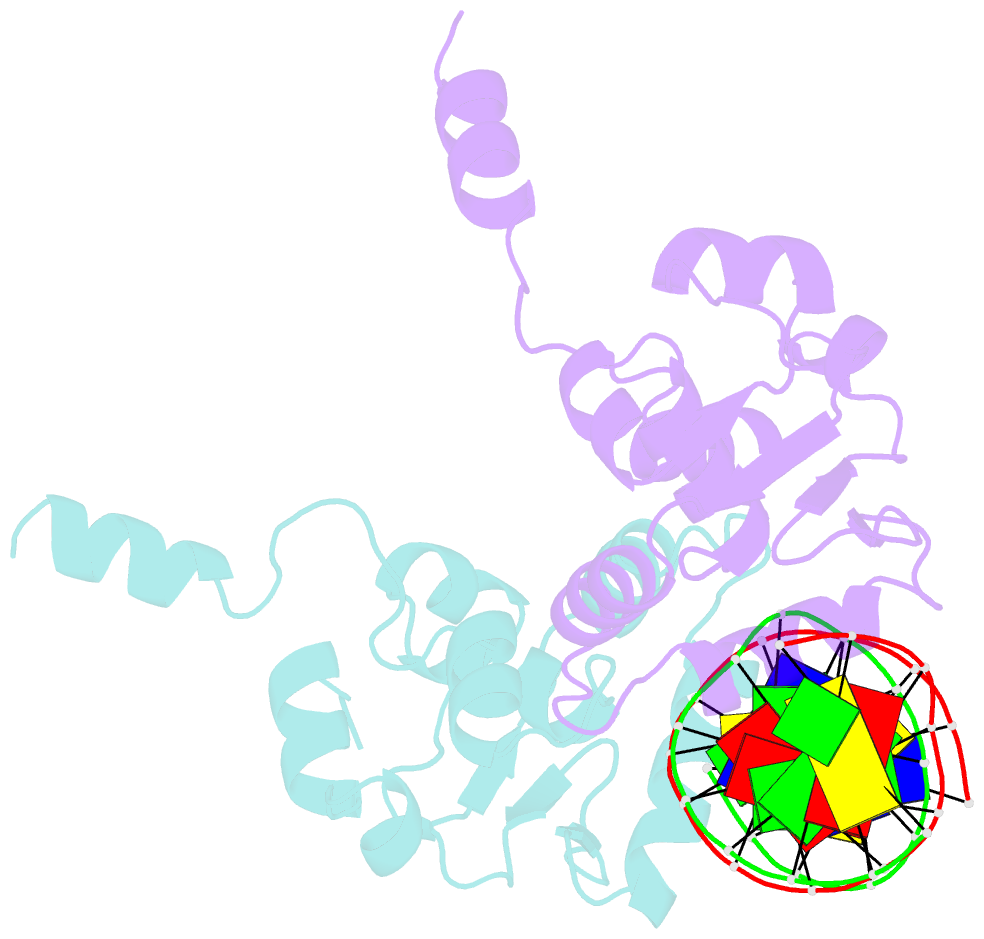
- Ets1 complex with stromelysin-1 promoter DNA
- Reference
- Babayeva ND, Wilder PJ, Shiina M, Mino K, Desler M, Ogata K, Rizzino A, Tahirov TH (2010): "Structural basis of Ets1 cooperative binding to palindromic sequences on stromelysin-1 promoter DNA." Cell Cycle, 9, 3054-3062.
- Abstract
- Ets1 is a member of the Ets family of transcription factors. Ets1 is autoinhibited and its activation requires heterodimerization with a partner protein or DNA-mediated homodimerization for cooperative DNA binding. In the latter case, Ets1 molecules bind to palindromic sequences in which two Ets-binding sites (EBS) are separated by four base pairs, for example in the promoters of stromelysin-1 and p53. Interestingly, counteraction of autoinhibition requires the autoinhibitory region encoded by exon VII of the gene. The structural basis for the requirement of autoinhibitory sequences for Ets1 binding to palindromic EBS still remains unresolved. Here we report the crystal structure of two Ets1 molecules bound to an EBS palindrome of the stromelysin-1 promoter DNA, providing a plausible explanation for the requirement of exon VII-encoded sequences for Ets1 cooperative DNA binding. The proposed mechanism was verified both in vitro by surface plasmon resonance and in vivo by transcription-based assays.