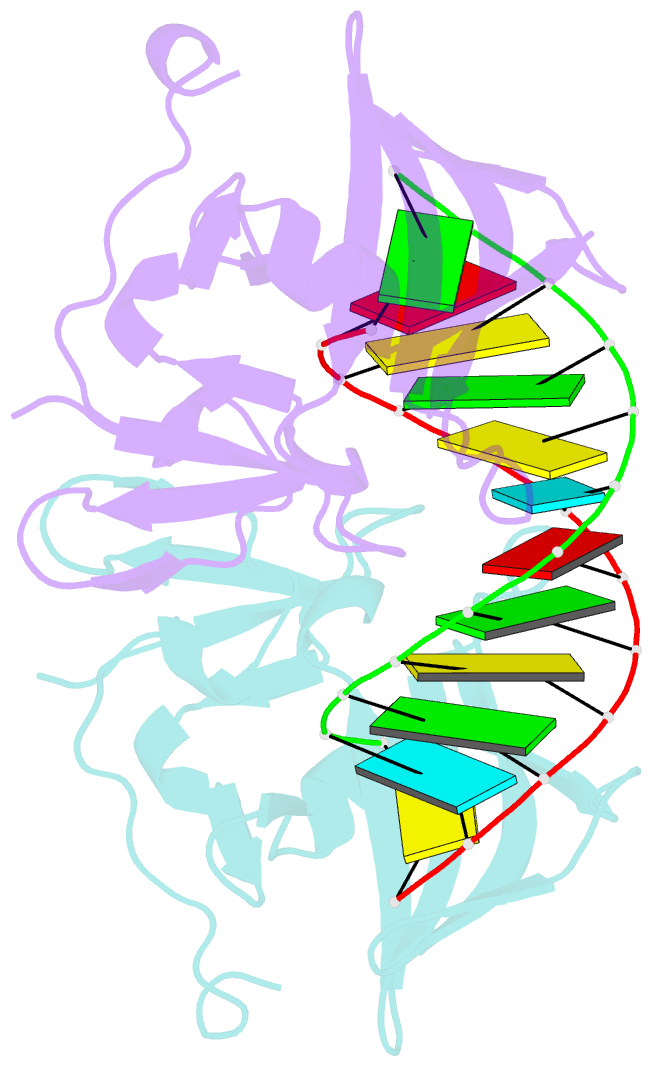
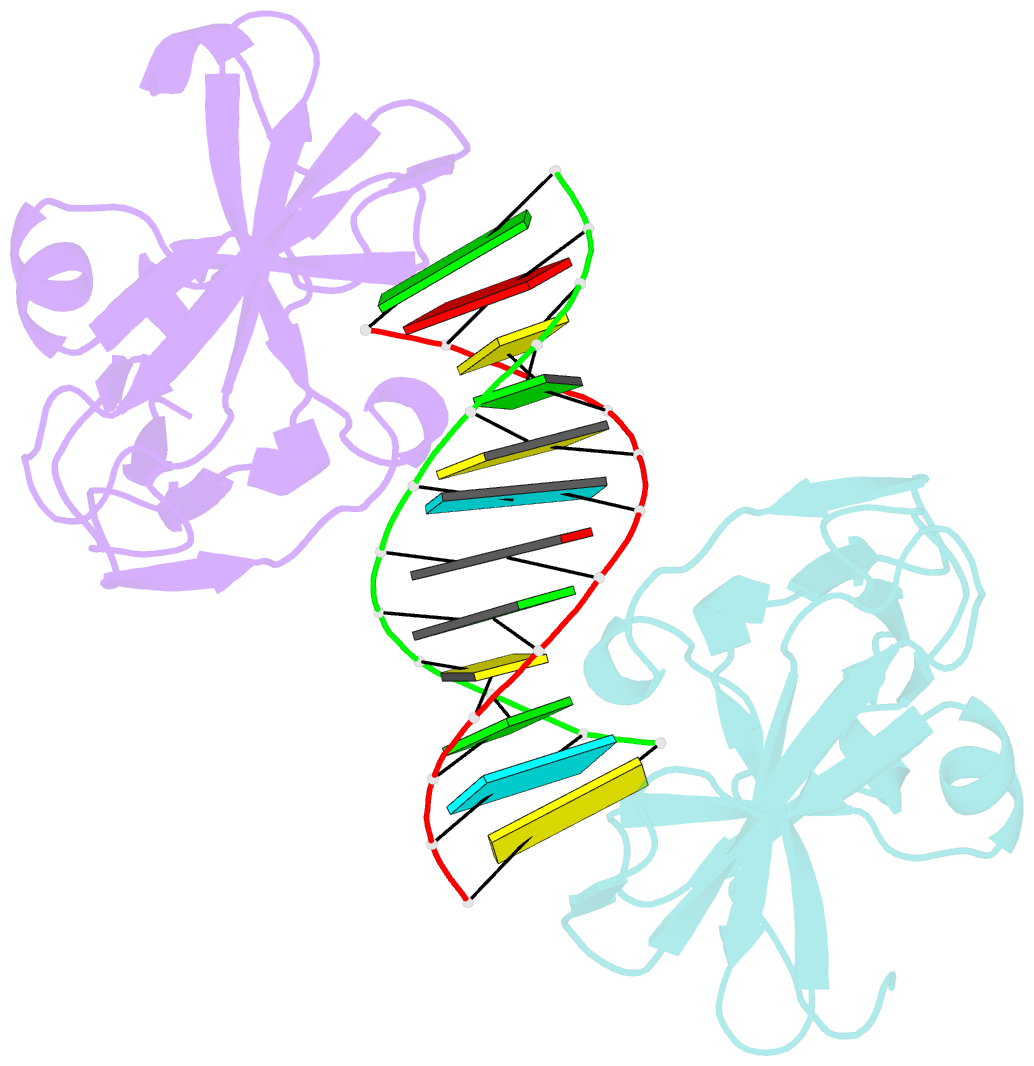
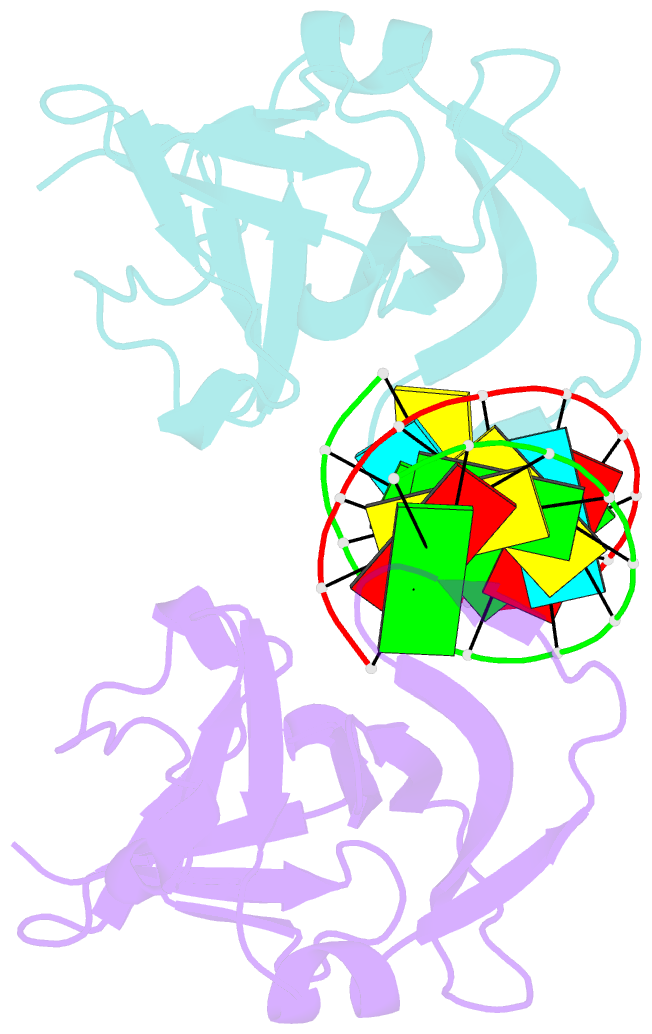
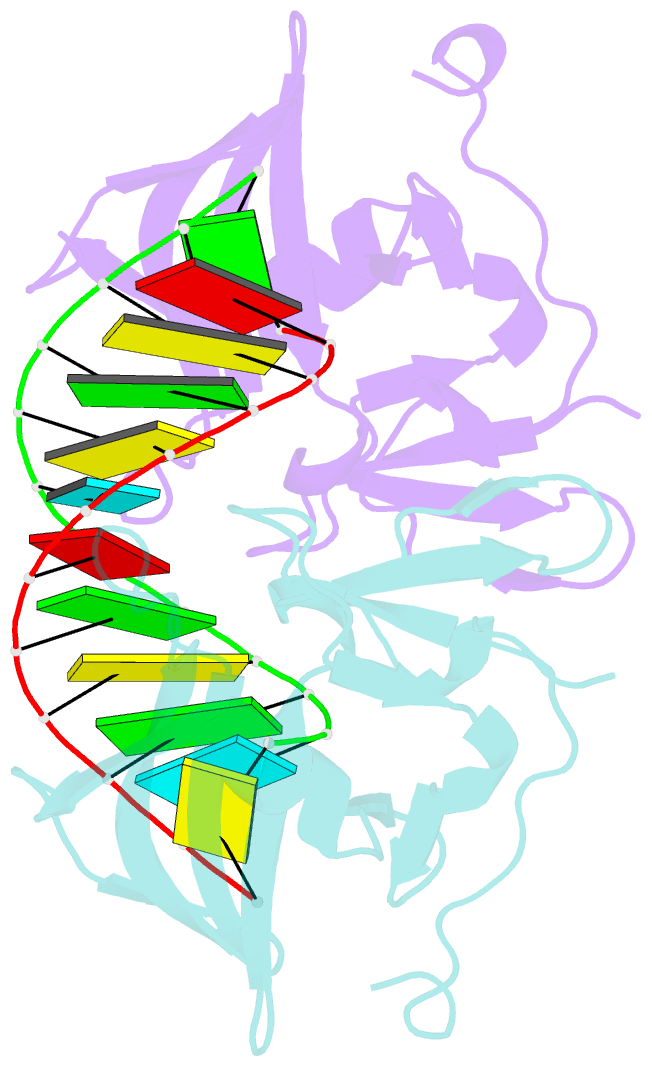
Summary information and primary citation
- PDB-id
- 3ncu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.55 Å)
- Summary
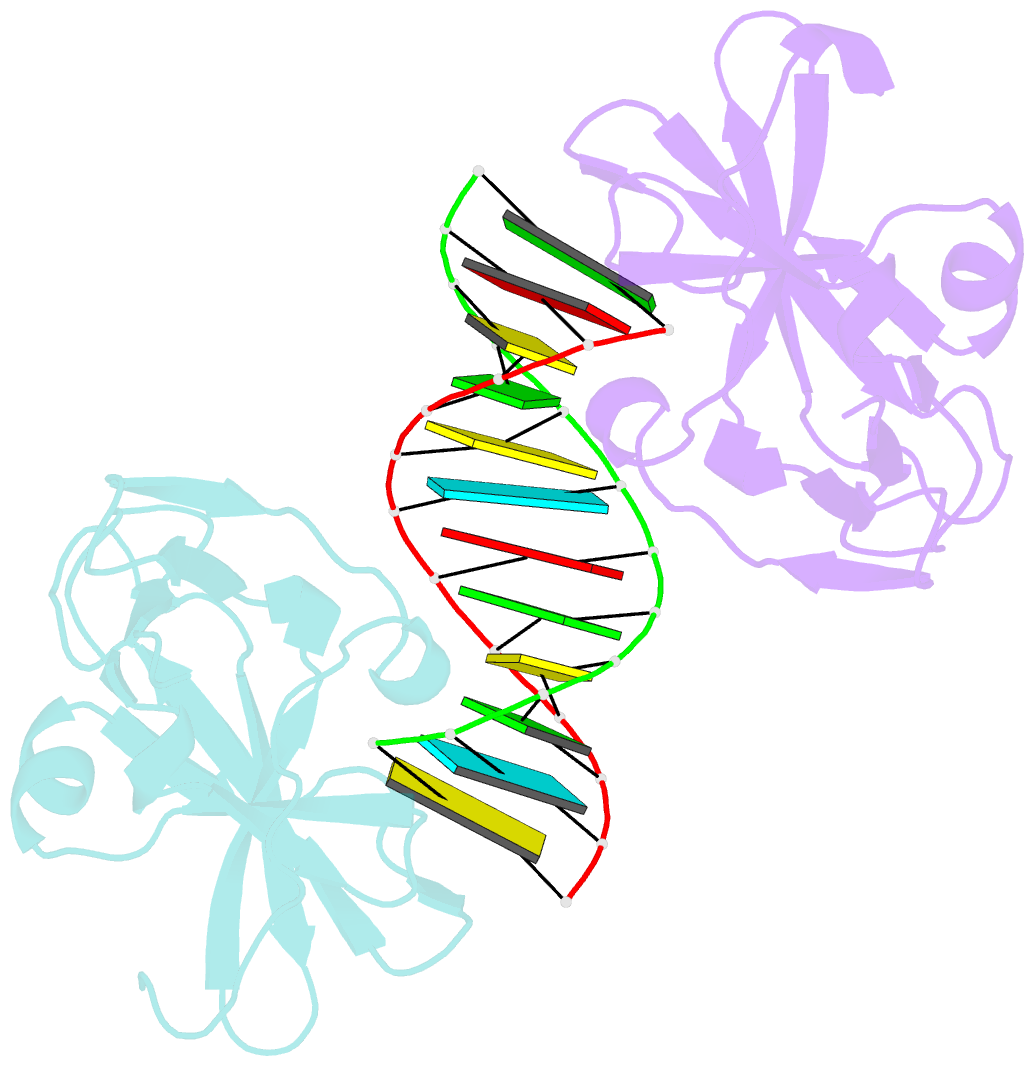
- Structural and functional insights into pattern recognition by the innate immune receptor rig-i
- Reference
- Wang Y, Ludwig J, Schuberth C, Goldeck M, Schlee M, Li H, Juranek S, Sheng G, Micura R, Tuschl T, Hartmann G, Patel DJ (2010): "Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I." Nat.Struct.Mol.Biol., 17, 781-787. doi: 10.1038/nsmb.1863.
- Abstract
- RIG-I is a cytosolic helicase that senses 5'-ppp RNA contained in negative-strand RNA viruses and triggers innate antiviral immune responses. Calorimetric binding studies established that the RIG-I C-terminal regulatory domain (CTD) binds to blunt-end double-stranded 5'-ppp RNA a factor of 17 more tightly than to its single-stranded counterpart. Here we report on the crystal structure of RIG-I CTD bound to both blunt ends of a self-complementary 5'-ppp dsRNA 12-mer, with interactions involving 5'-pp clearly visible in the complex. The structure, supported by mutation studies, defines how a lysine-rich basic cleft within the RIG-I CTD sequesters the observable 5'-pp of the bound RNA, with a stacked phenylalanine capping the terminal base pair. Key intermolecular interactions observed in the crystalline state are retained in the complex of 5'-ppp dsRNA 24-mer and full-length RIG-I under in vivo conditions, as evaluated from the impact of binding pocket RIG-I mutations and 2'-OCH(3) RNA modifications on the interferon response.