Summary information and primary citation
- PDB-id
- 3ndb; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- signaling protein-RNA
- Method
- X-ray (3.0 Å)
- Summary
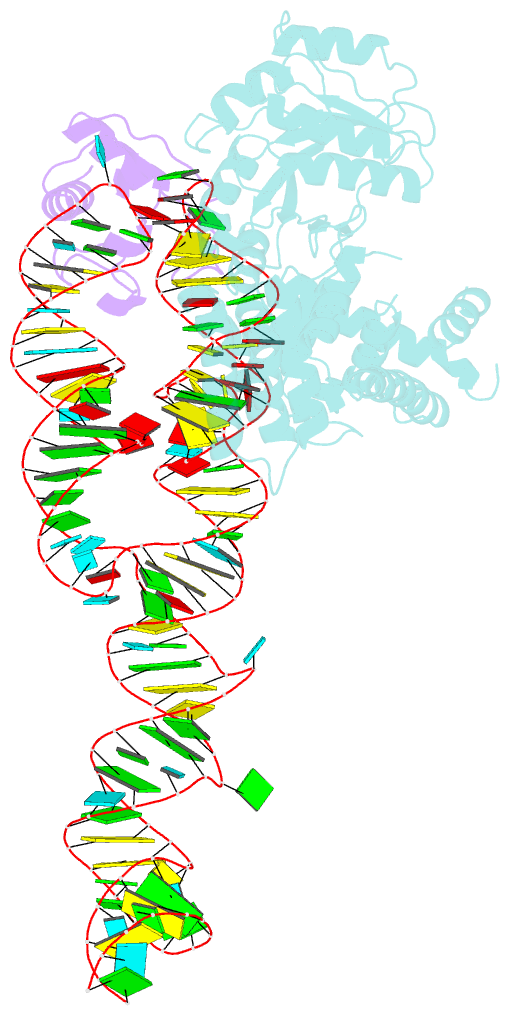
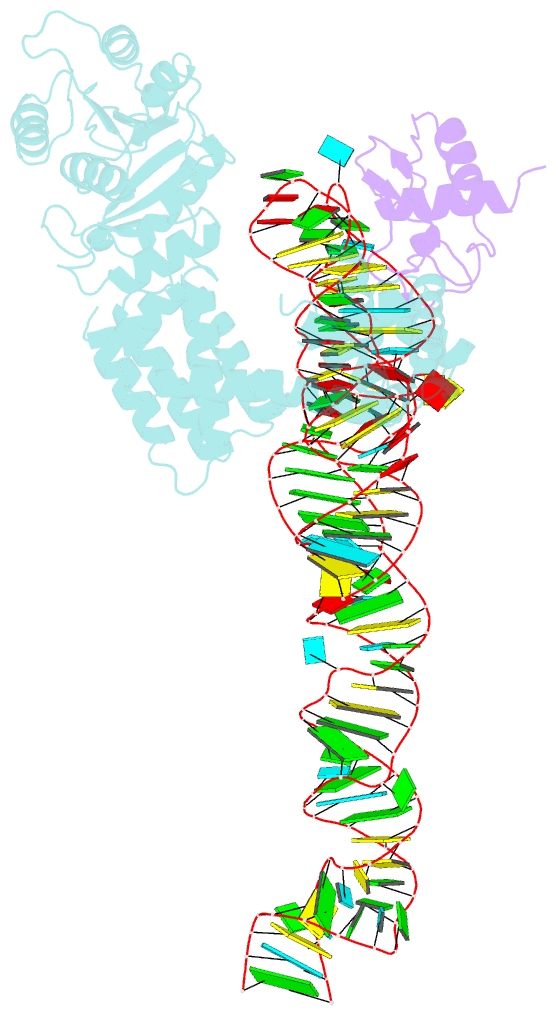
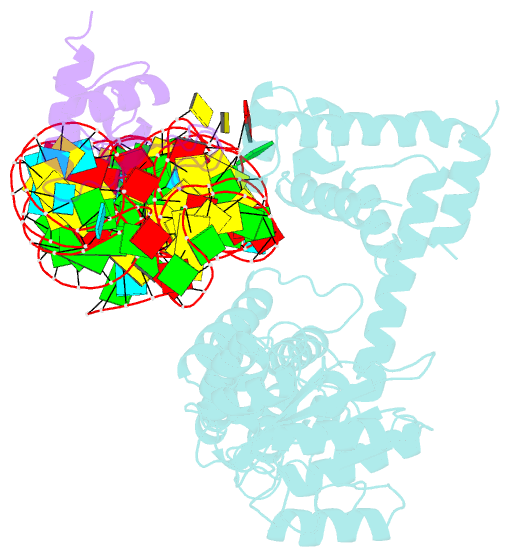
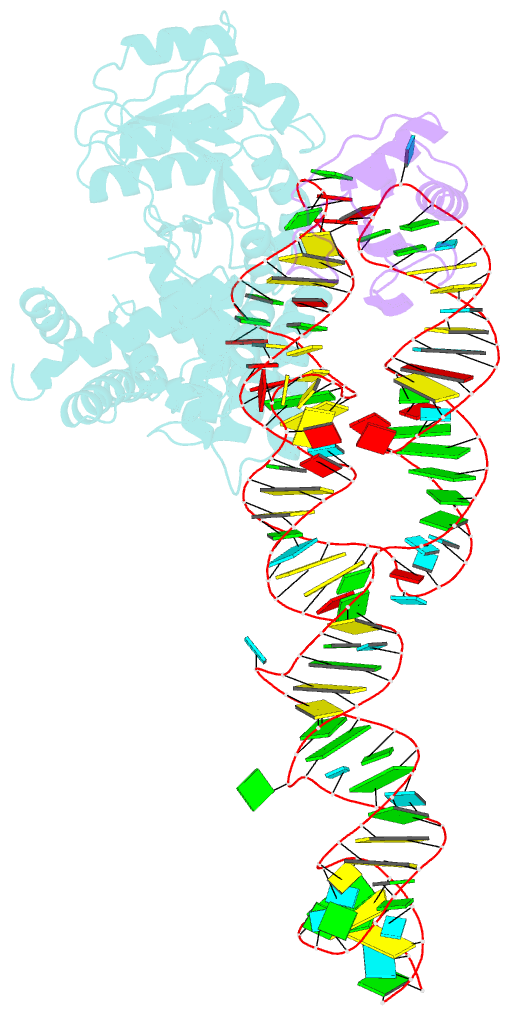
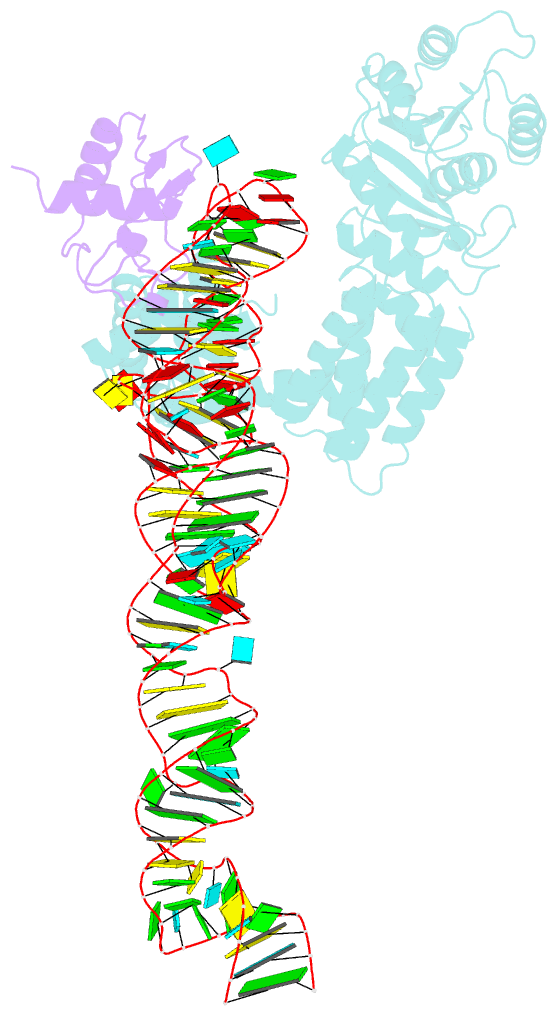
- Crystal structure of a signal sequence bound to the signal recognition particle
- Reference
- Hainzl T, Huang S, Merilainen G, Brannstrom K, Sauer-Eriksson AE (2011): "Structural basis of signal-sequence recognition by the signal recognition particle." Nat.Struct.Mol.Biol., 18, 389-391. doi: 10.1038/nsmb.1994.
- Abstract
- The signal recognition particle (SRP) recognizes and binds the signal sequence of nascent proteins as they emerge from the ribosome. We present here the 3.0-Å structure of a signal sequence bound to the Methanococcus jannaschii SRP core. Structural comparison with the free SRP core shows that signal-sequence binding induces formation of the GM-linker helix and a 180° flip of the NG domain-structural changes that ensure a hierarchical succession of events during protein targeting.