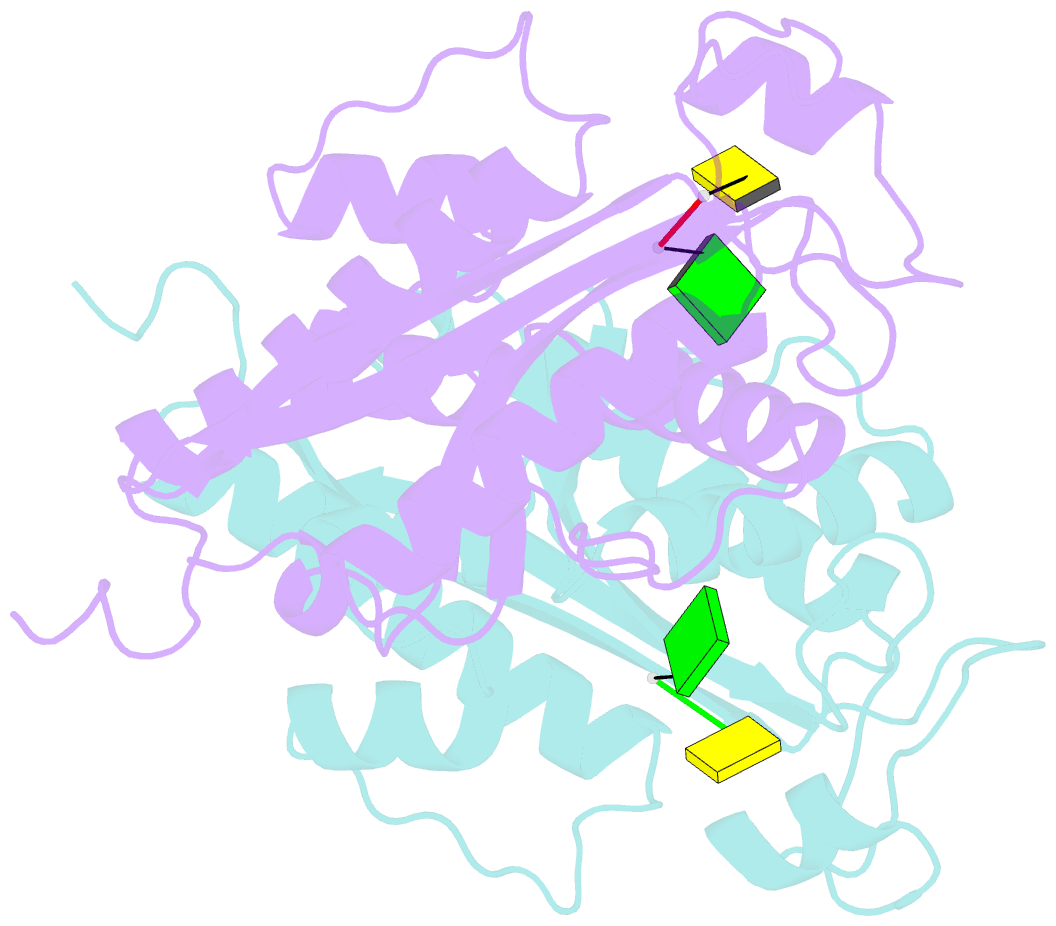
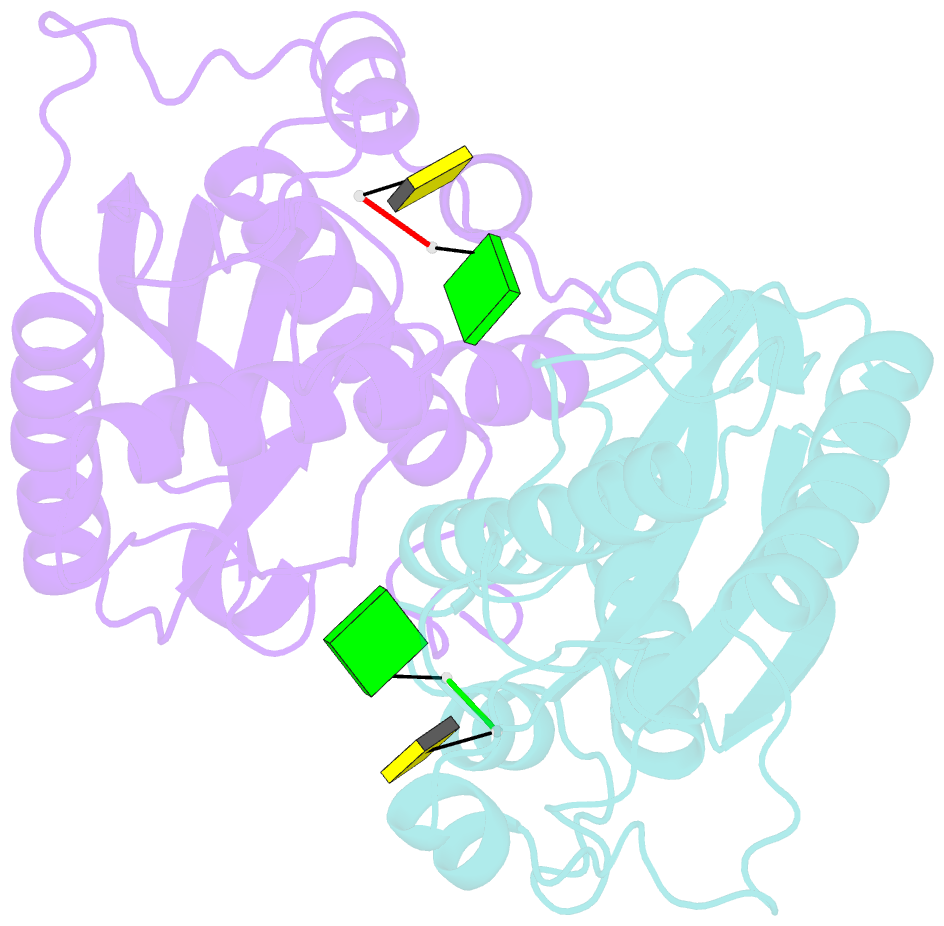
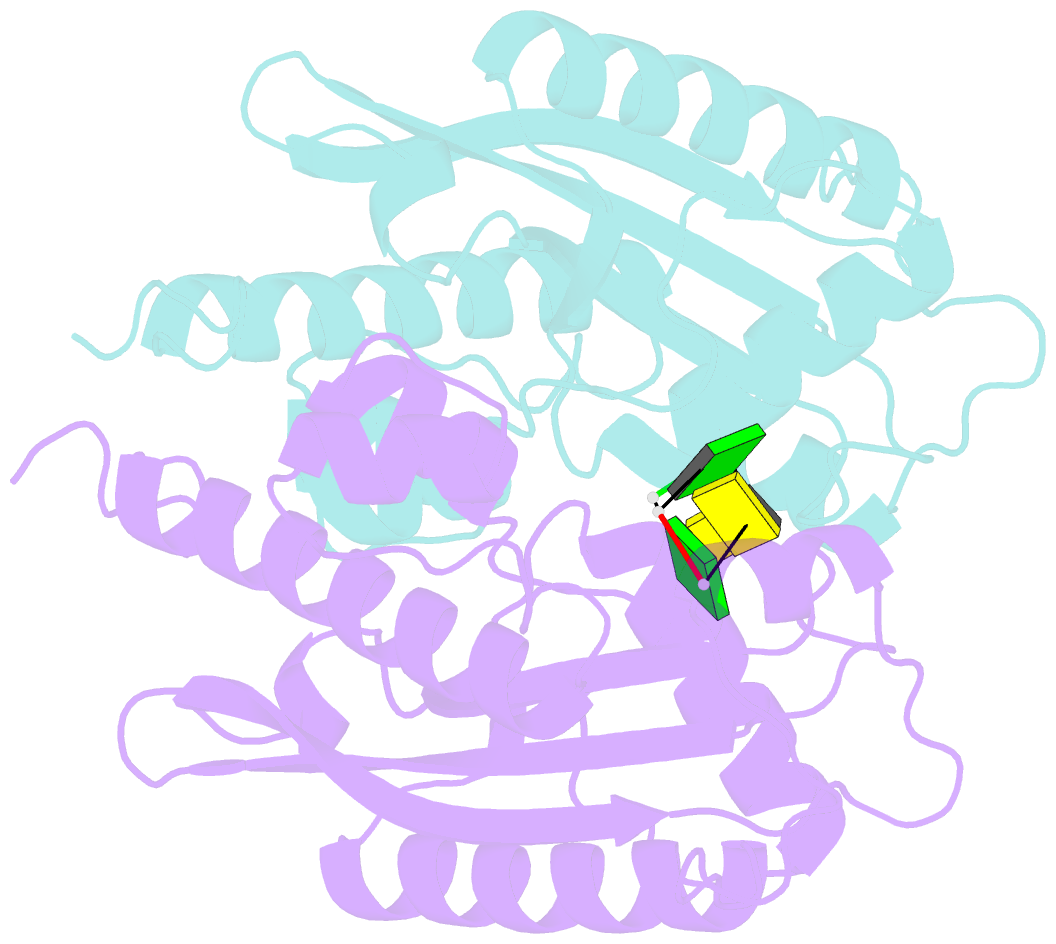
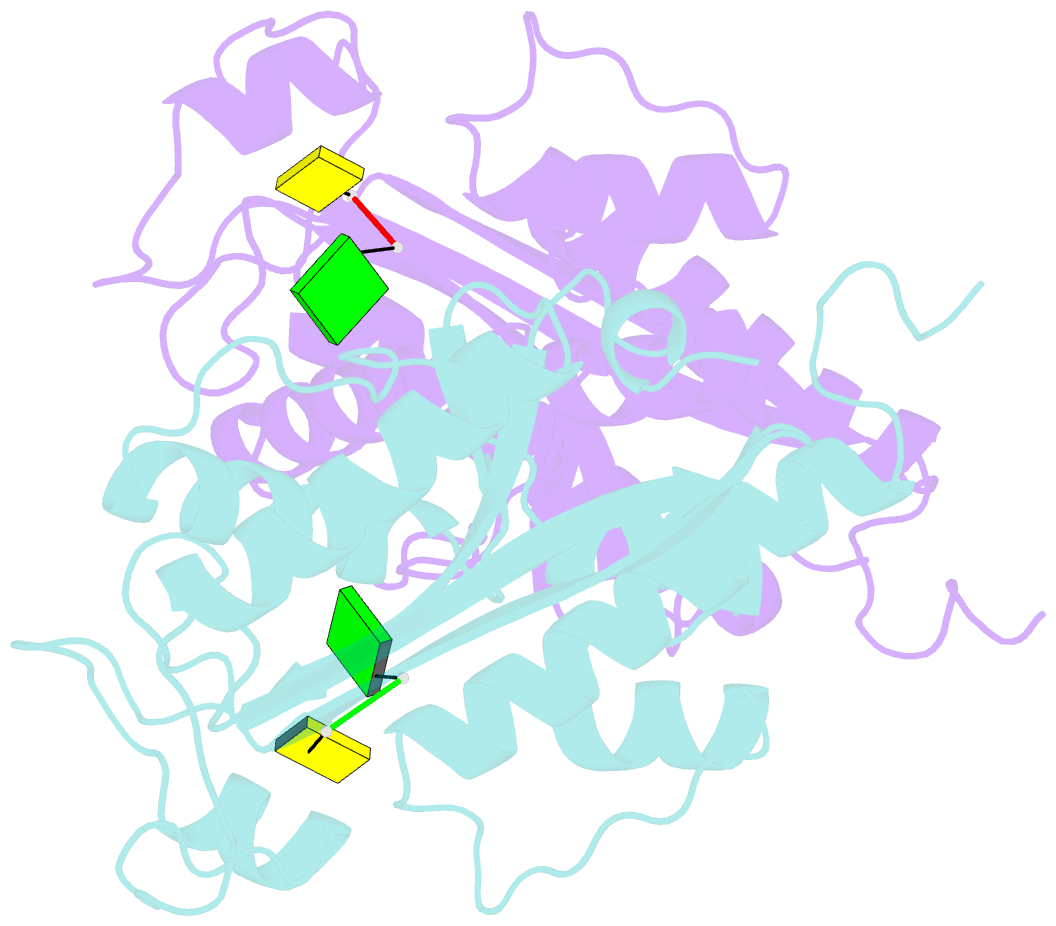
Summary information and primary citation
- PDB-id
- 3ngz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.1 Å)
- Summary
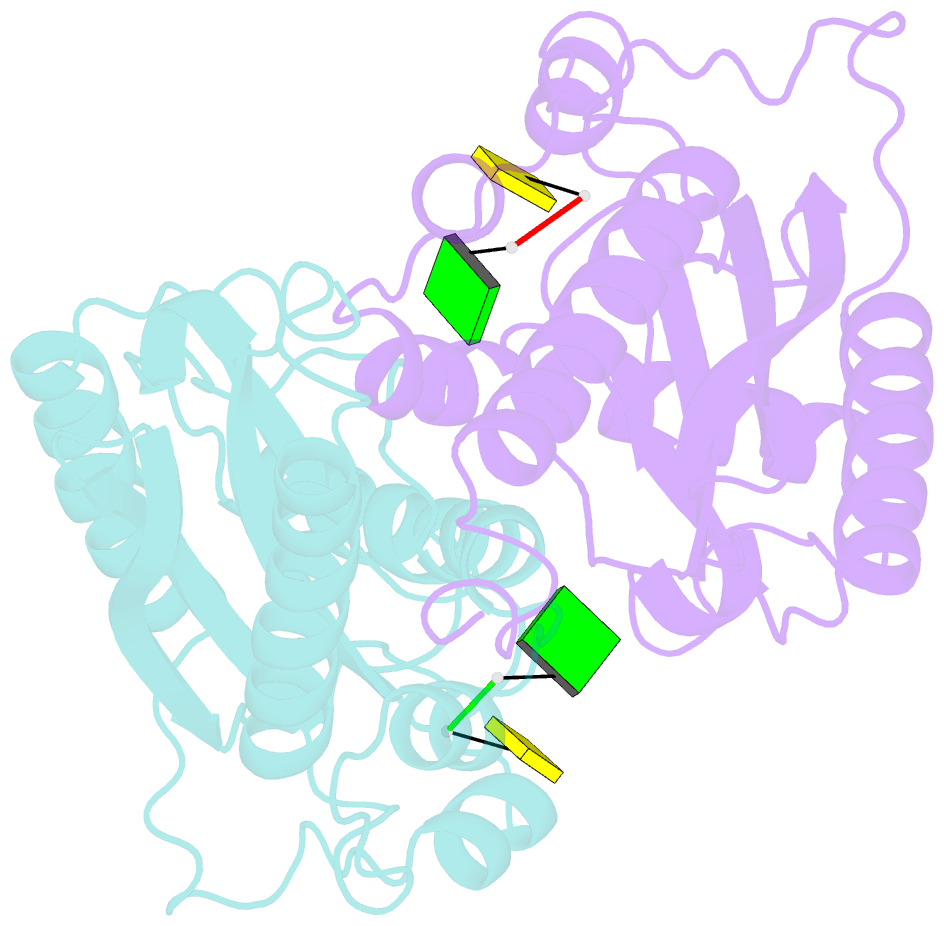
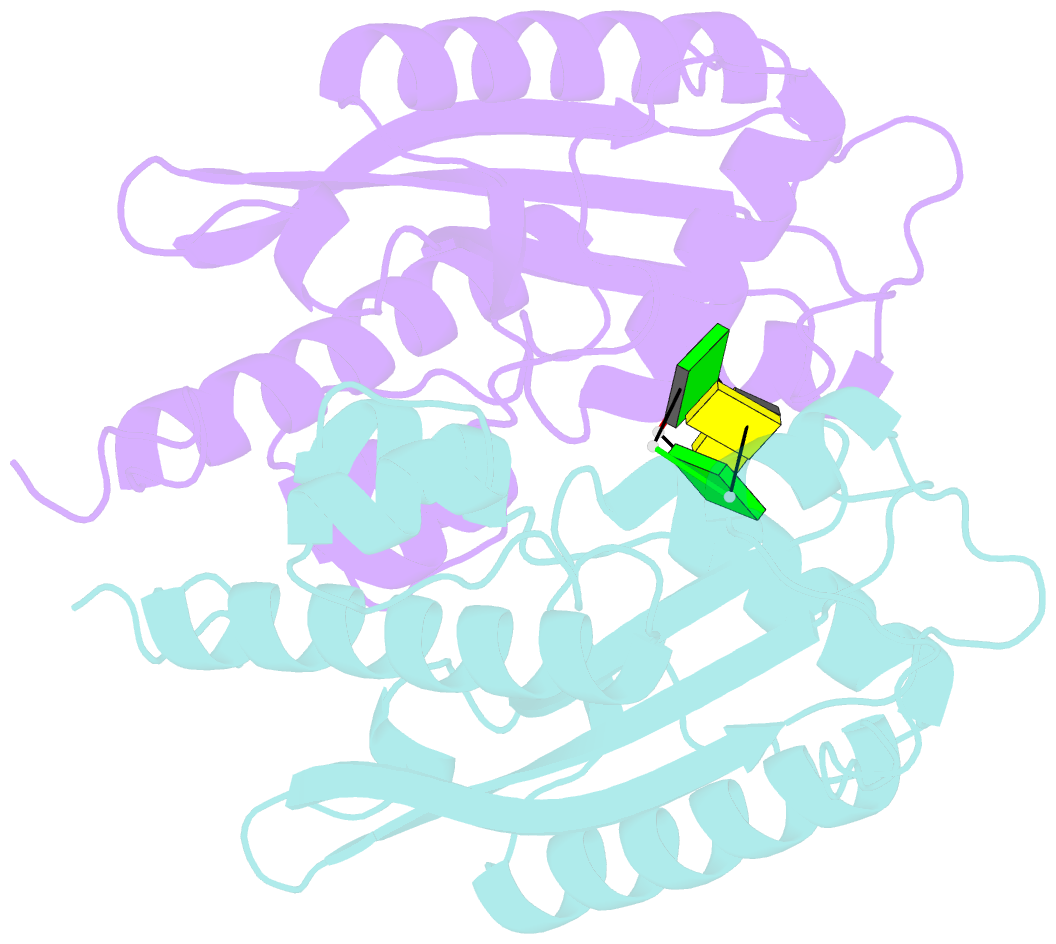
- Crystal structure of rnase t in complex with a non-preferred ssDNA (gc) with one mg in the active site
- Reference
- Hsiao Y-Y, Yang C-C, Lin CL, Lin JLJ, Duh Y, Yuan HS (2011): "Structural basis for RNA trimming by RNase T in stable RNA 3'-end maturation." Nat.Chem.Biol., 7, 236-243. doi: 10.1038/nchembio.524.
- Abstract
- RNA maturation relies on various exonucleases to remove nucleotides successively from the 5' or 3' end of nucleic acids. However, little is known regarding the molecular basis for substrate and cleavage preference of exonucleases. Our biochemical and structural analyses on RNase T-DNA complexes show that the RNase T dimer has an ideal architecture for binding a duplex with a short 3' overhang to produce a digestion product of a duplex with a 2-nucleotide (nt) or 1-nt 3' overhang, depending on the composition of the last base pair in the duplex. A 'C-filter' in RNase T screens out the nucleic acids with 3'-terminal cytosines for hydrolysis by inducing a disruptive conformational change at the active site. Our results reveal the general principles and the working mechanism for the final trimming step made by RNase T in the maturation of stable RNA and pave the way for the understanding of other DEDD family exonucleases.