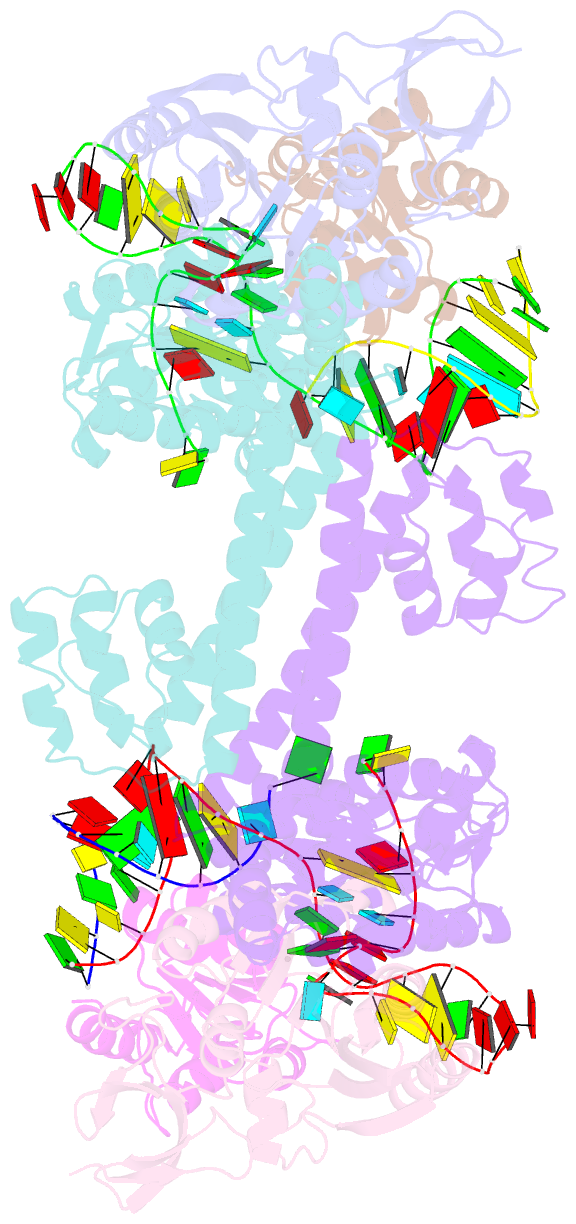
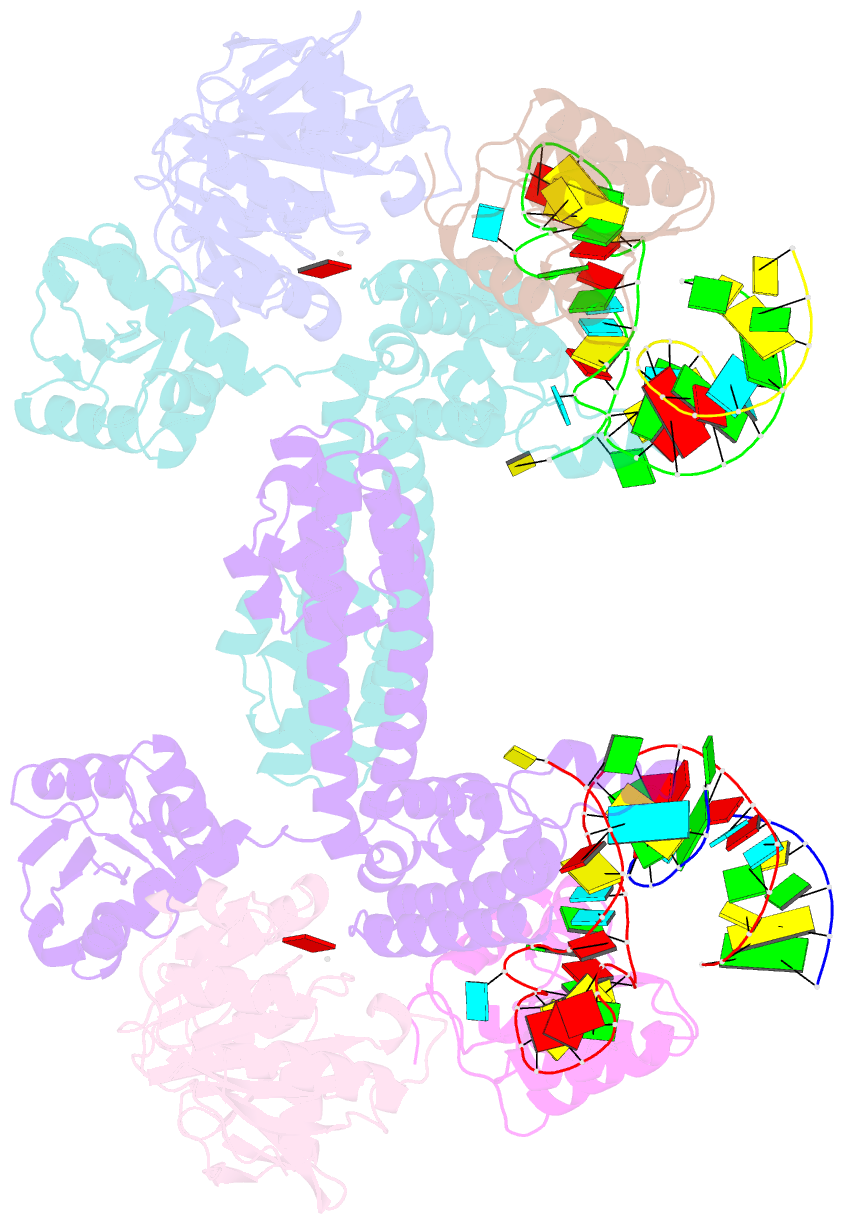
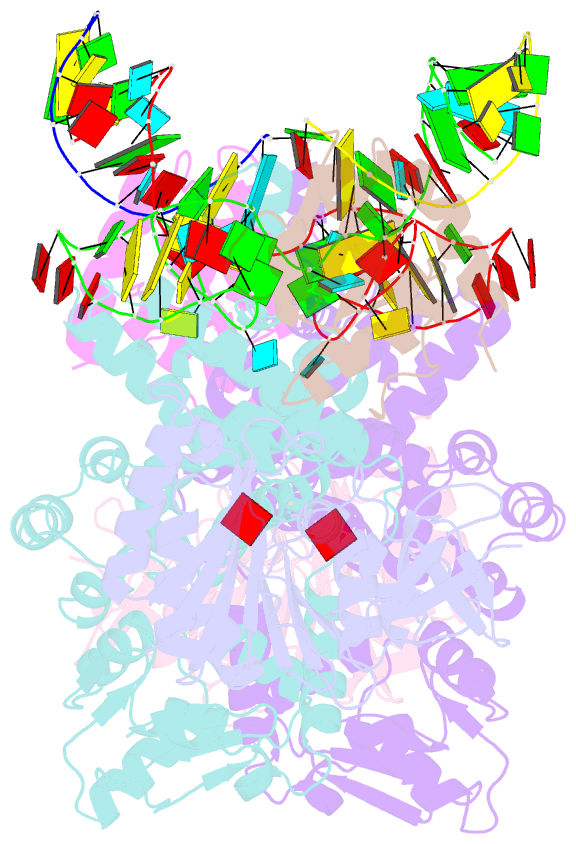
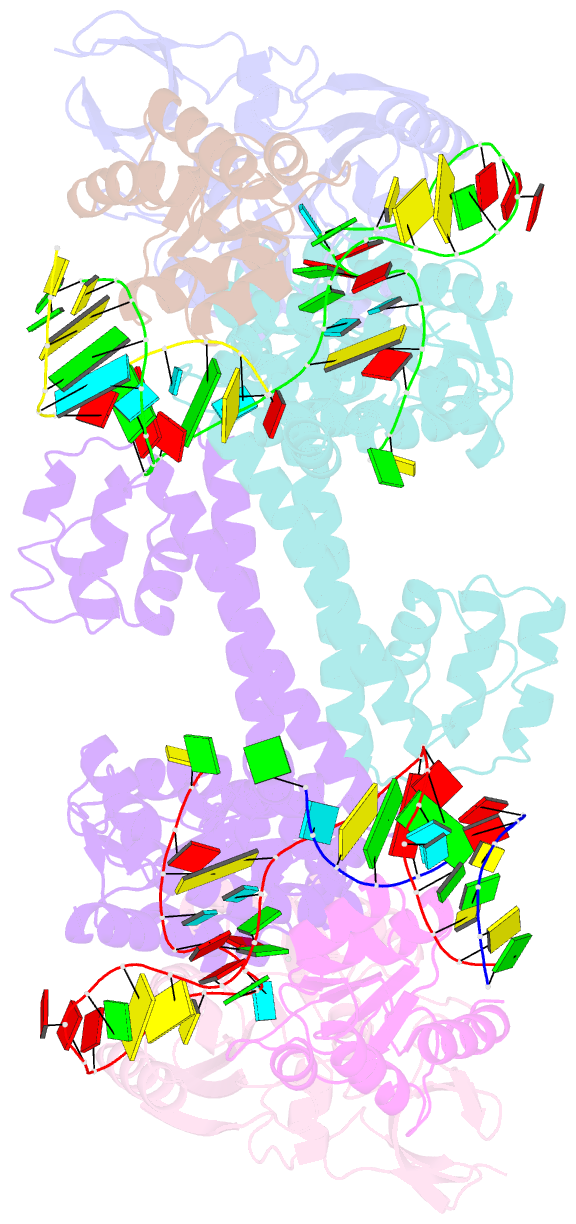
Summary information and primary citation
- PDB-id
- 3nmu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.729 Å)
- Summary
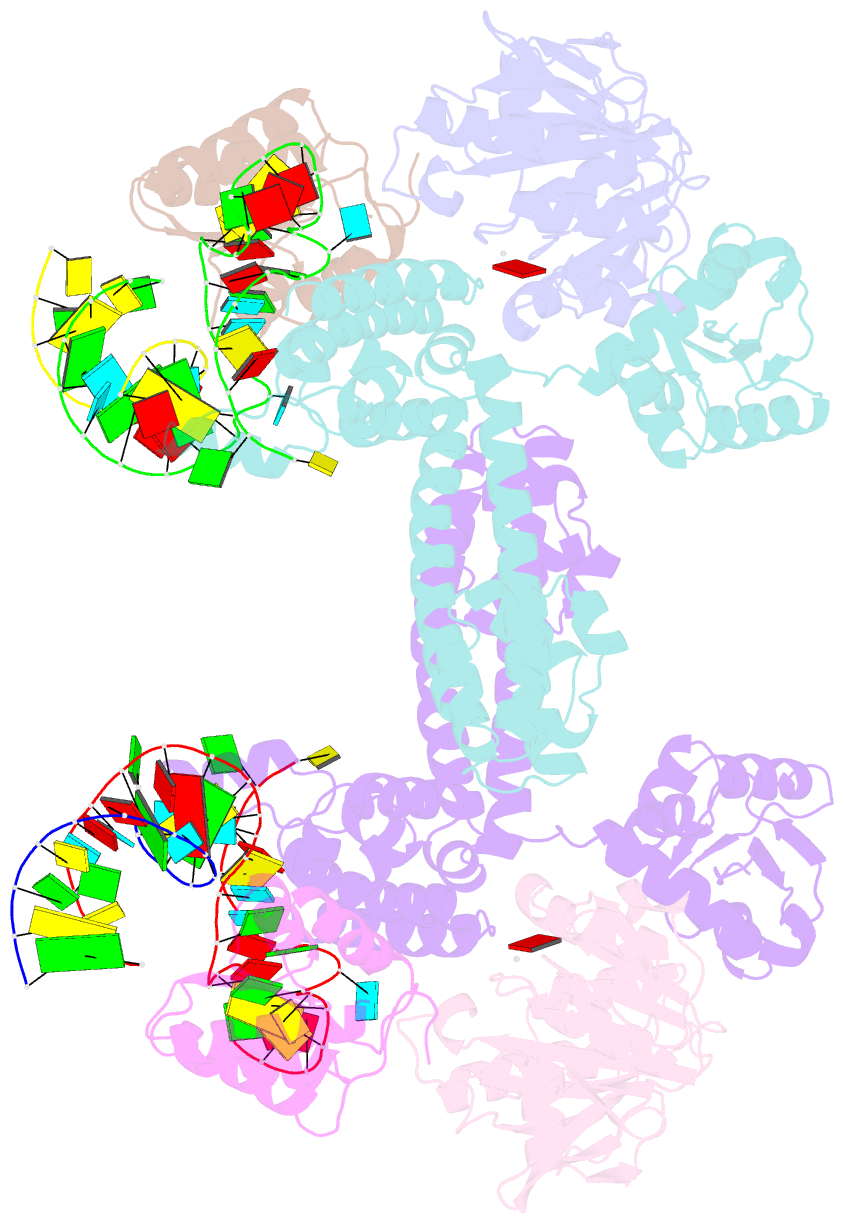
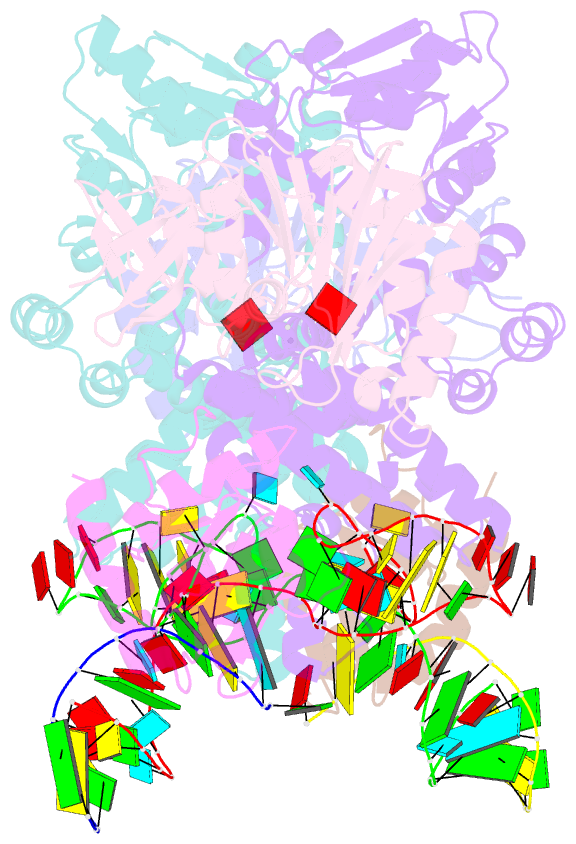
- Crystal structure of substrate-bound halfmer box c-d rnp
- Reference
- Xue S, Wang R, Yang F, Terns RM, Terns MP, Zhang X, Maxwell ES, Li H (2010): "Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle." Mol.Cell, 39, 939-949. doi: 10.1016/j.molcel.2010.08.022.
- Abstract
- Box C/D small nucleolar and Cajal body ribonucleoprotein particles (sno/scaRNPs) direct site-specific 2'-O-methylation of ribosomal and spliceosomal RNAs and are critical for gene expression. Here we report crystal structures of an archaeal box C/D RNP containing three core proteins (fibrillarin, Nop56/58, and L7Ae) and a half-mer box C/D guide RNA paired with a substrate RNA. The structure reveals a guide-substrate RNA duplex orientation imposed by a composite protein surface and the conserved GAEK motif of Nop56/58. Molecular modeling supports a dual C/D RNP structure that closely mimics that recently visualized by electron microscopy. The substrate-bound dual RNP model predicts an asymmetric protein distribution between the RNP that binds and methylates the substrate RNA. The predicted asymmetric nature of the holoenzyme is consistent with previous biochemical data on RNP assembly and provides a simple solution for accommodating base-pairing between the C/D guide RNA and large ribosomal and spliceosomal substrate RNAs.