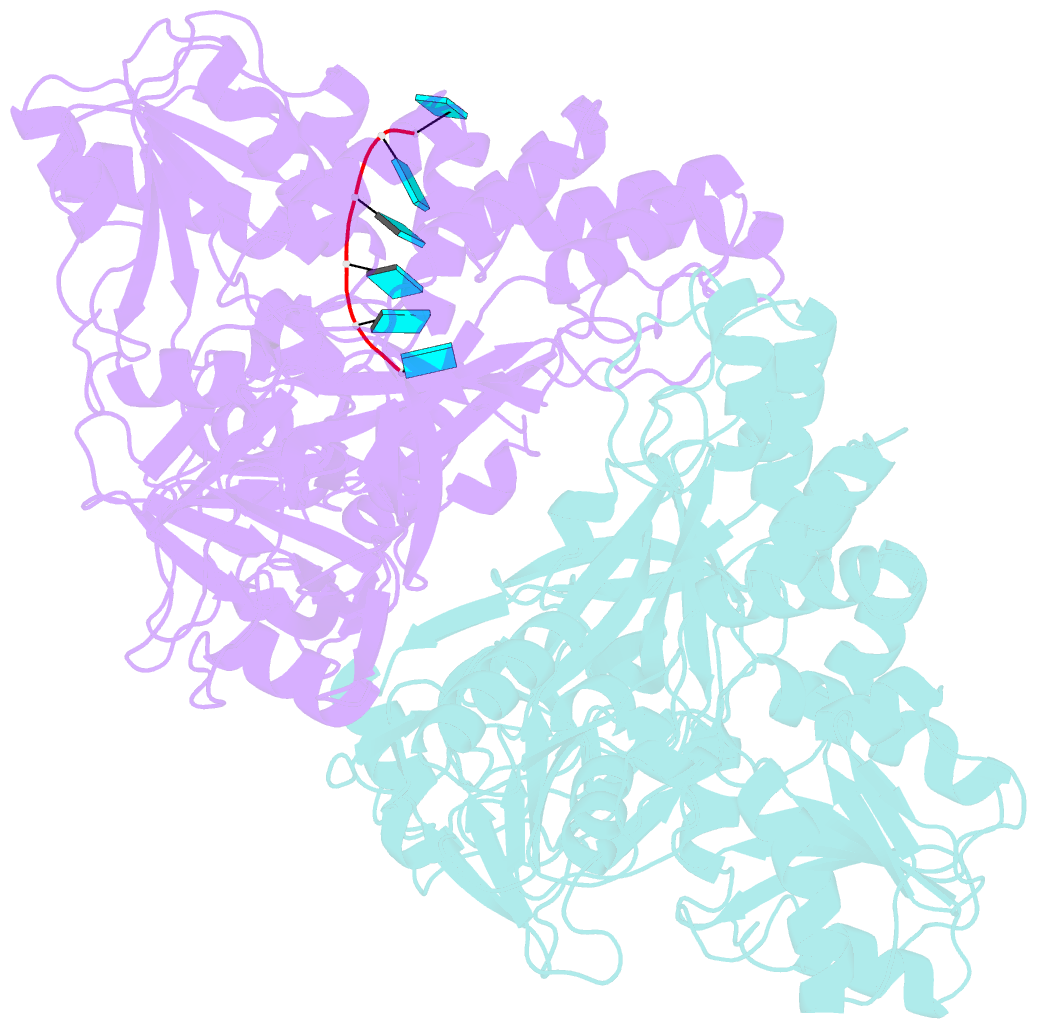
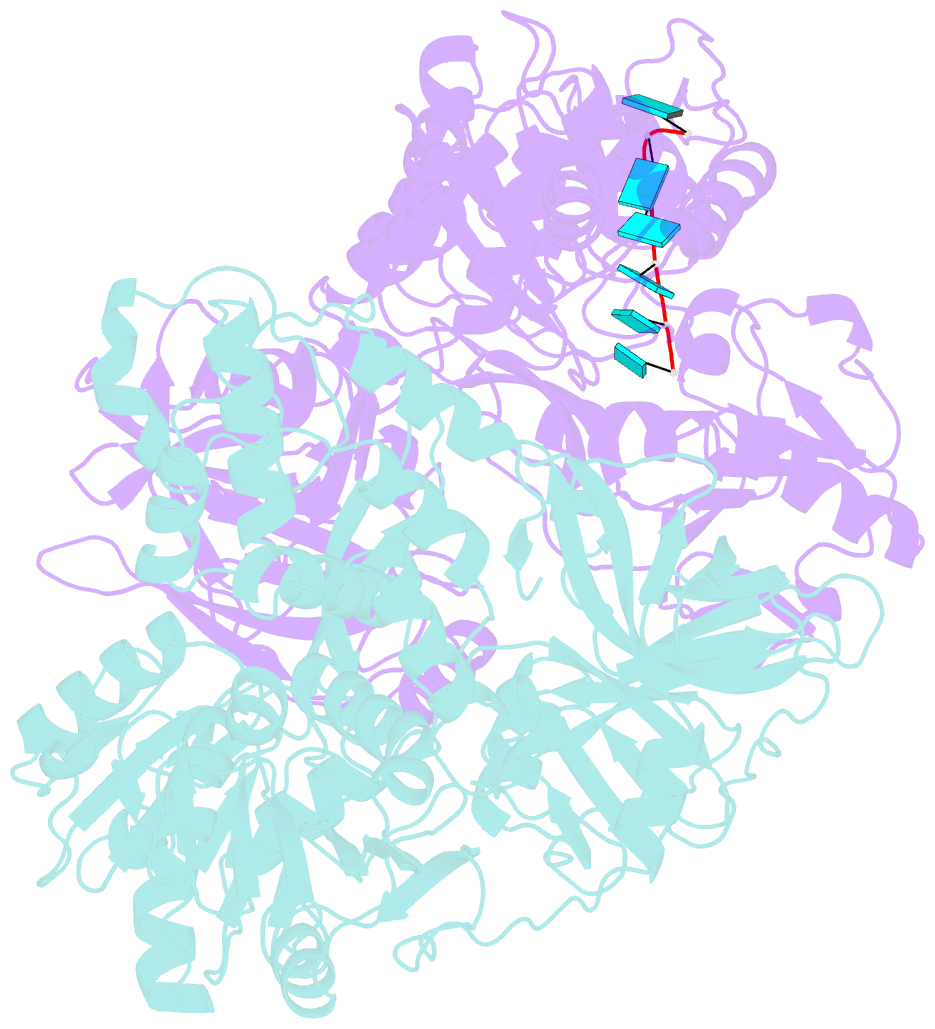
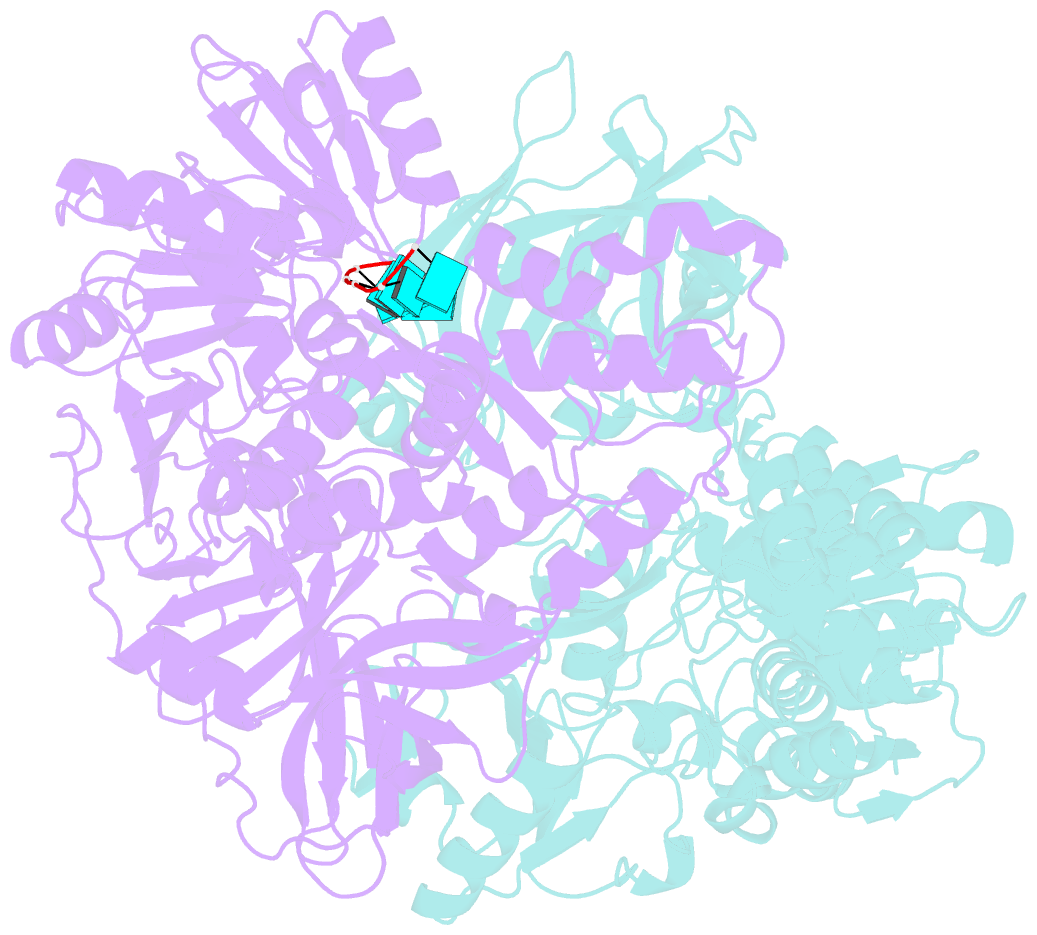
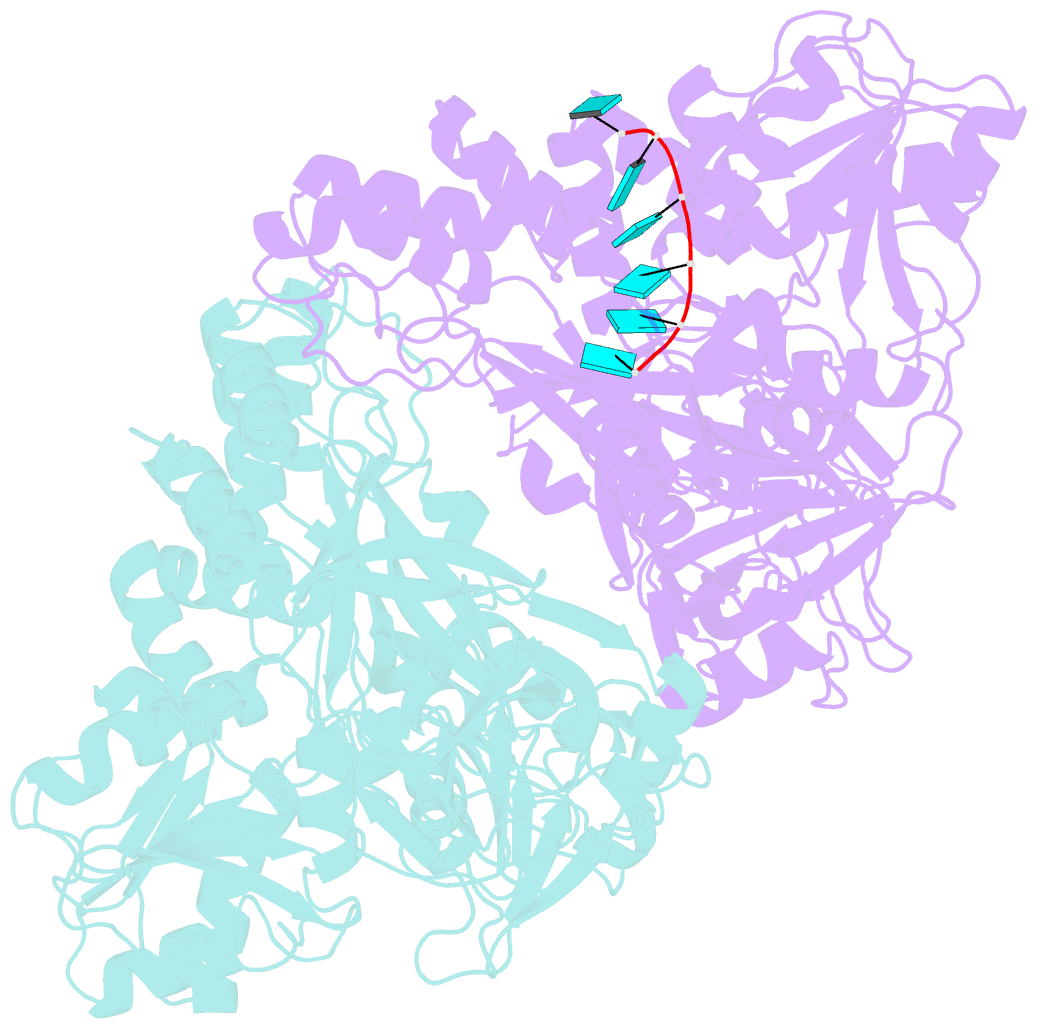
Summary information and primary citation
- PDB-id
- 3o8c; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.0 Å)
- Summary
- Visualizing atp-dependent RNA translocation by the ns3 helicase from hcv
- Reference
- Appleby TC, Anderson R, Fedorova O, Pyle AM, Wang R, Liu X, Brendza KM, Somoza JR (2011): "Visualizing ATP-Dependent RNA Translocation by the NS3 Helicase from HCV." J.Mol.Biol., 405, 1139-1153. doi: 10.1016/j.jmb.2010.11.034.
- Abstract
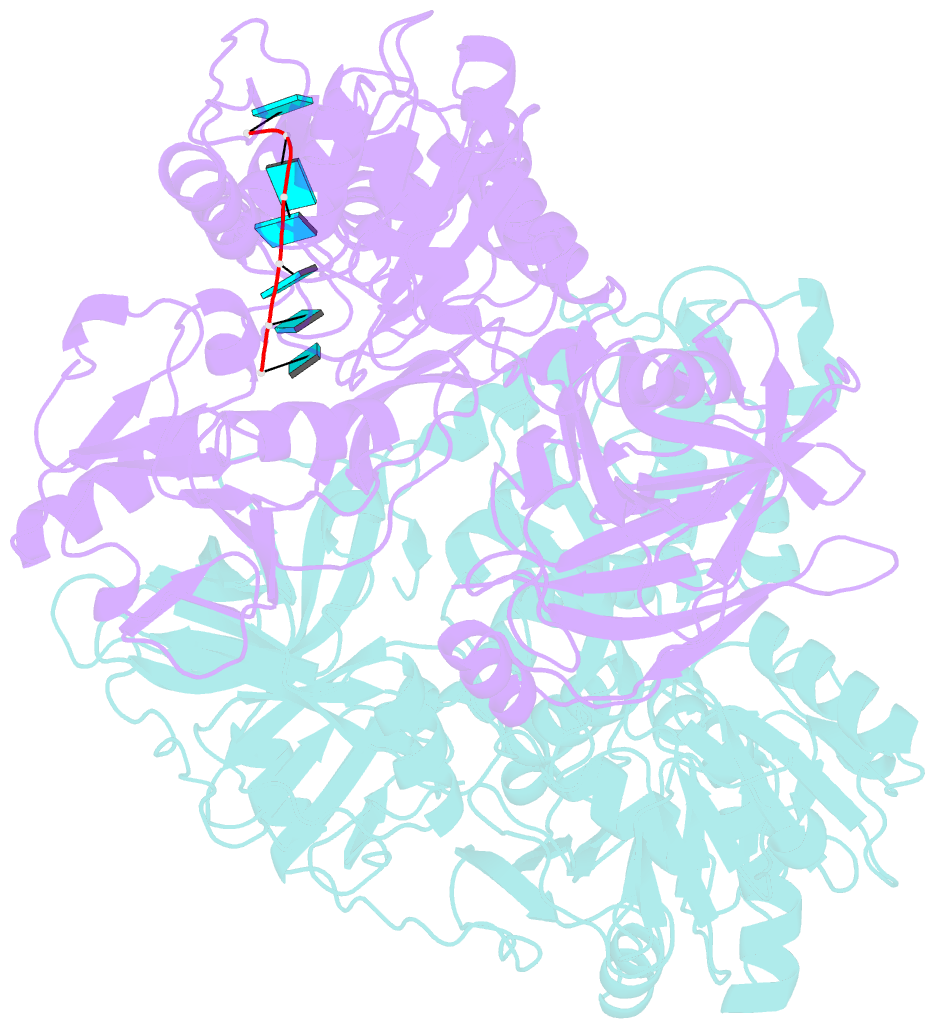
- The structural mechanism by which nonstructural protein 3 (NS3) from the hepatitis C virus (HCV) translocates along RNA is currently unknown. HCV NS3 is an ATP-dependent motor protein essential for viral replication and a member of the superfamily 2 helicases. Crystallographic analysis using a labeled RNA oligonucleotide allowed us to unambiguously track the positional changes of RNA bound to full-length HCV NS3 during two discrete steps of the ATP hydrolytic cycle. The crystal structures of HCV NS3, NS3 bound to bromine-labeled RNA, and a tertiary complex of NS3 bound to labeled RNA and a non-hydrolyzable ATP analog provide a direct view of how large domain movements resulting from ATP binding and hydrolysis allow the enzyme to translocate along the phosphodiester backbone. While directional translocation of HCV NS3 by a single base pair per ATP hydrolyzed is observed, the 3' end of the RNA does not shift register with respect to a conserved tryptophan residue, supporting a "spring-loading" mechanism that leads to larger steps by the enzyme as it moves along a nucleic acid substrate.