Summary information and primary citation
- PDB-id
- 3oha; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.0 Å)
- Summary
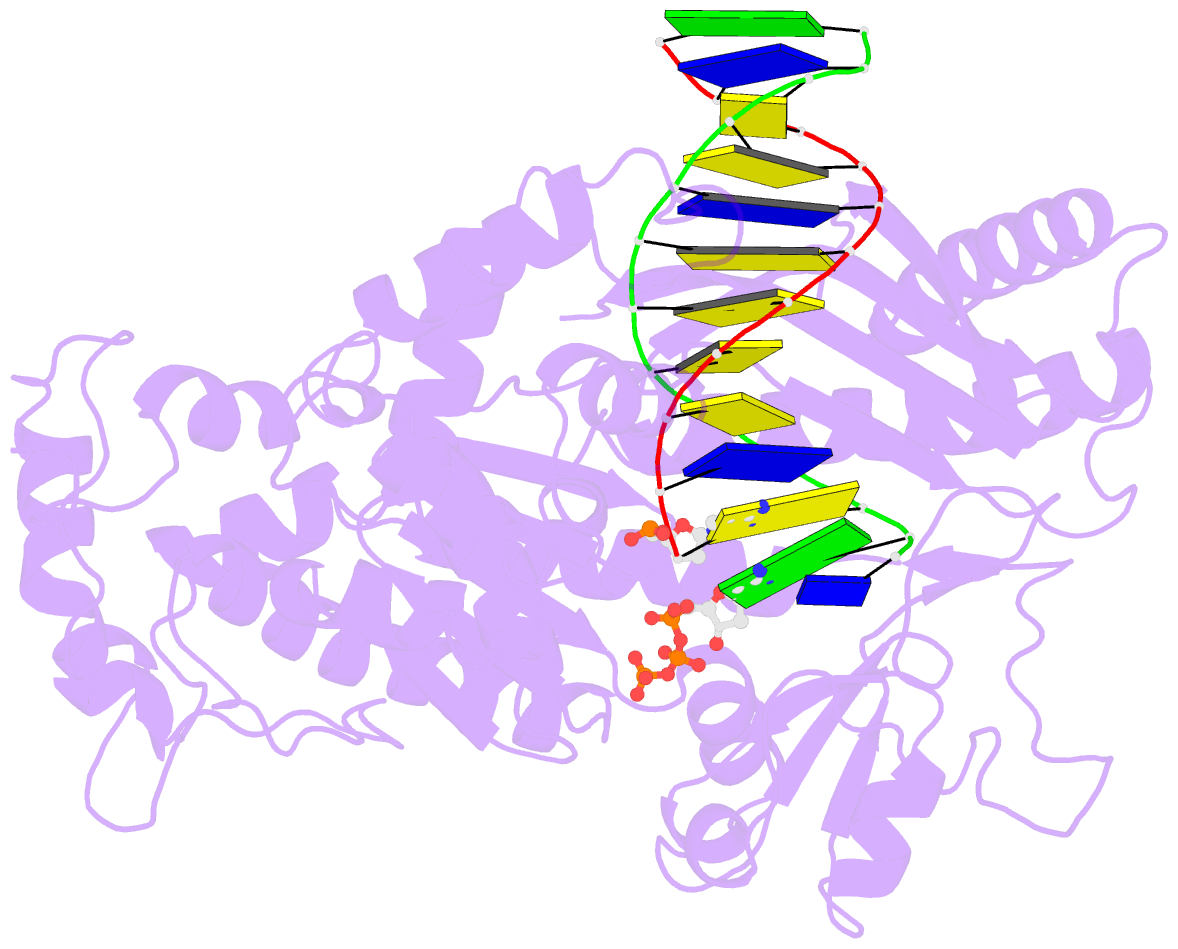
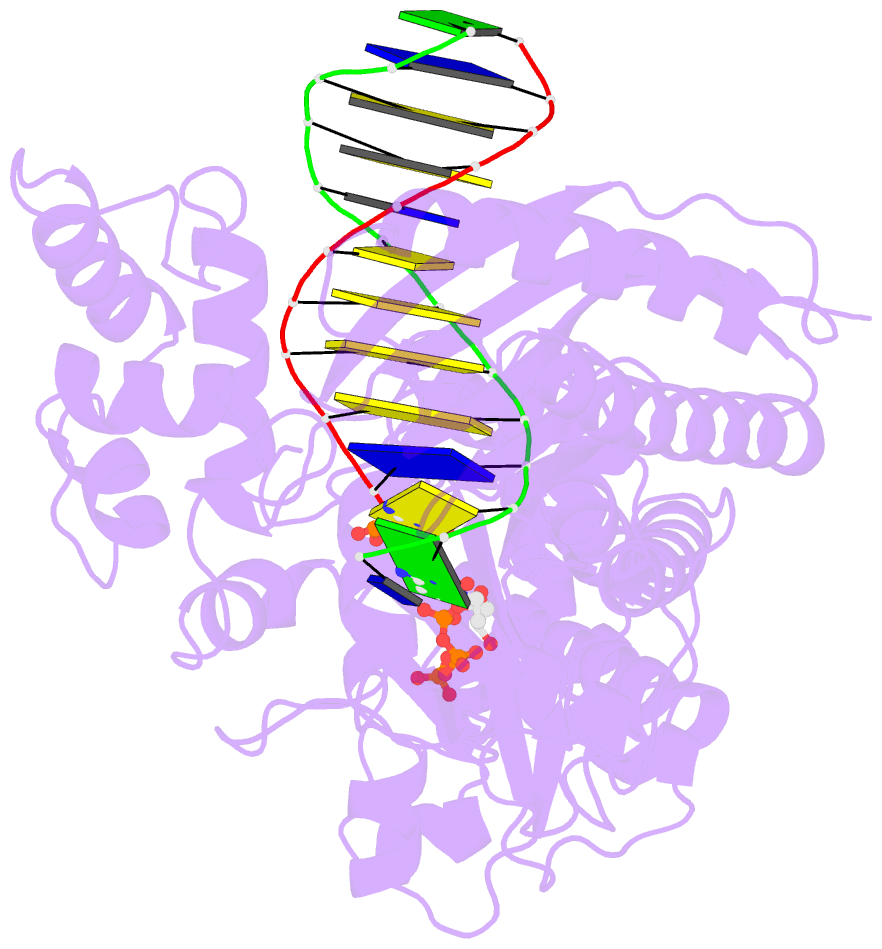
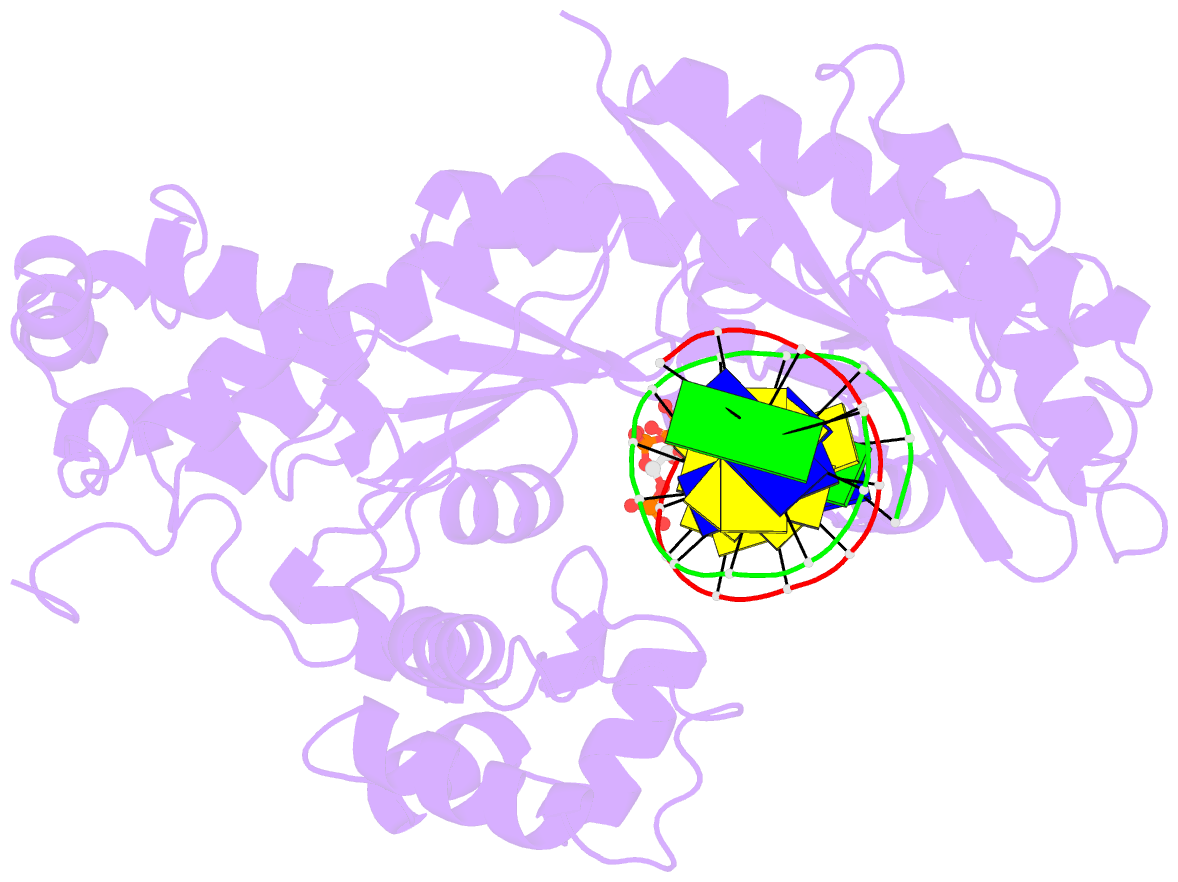
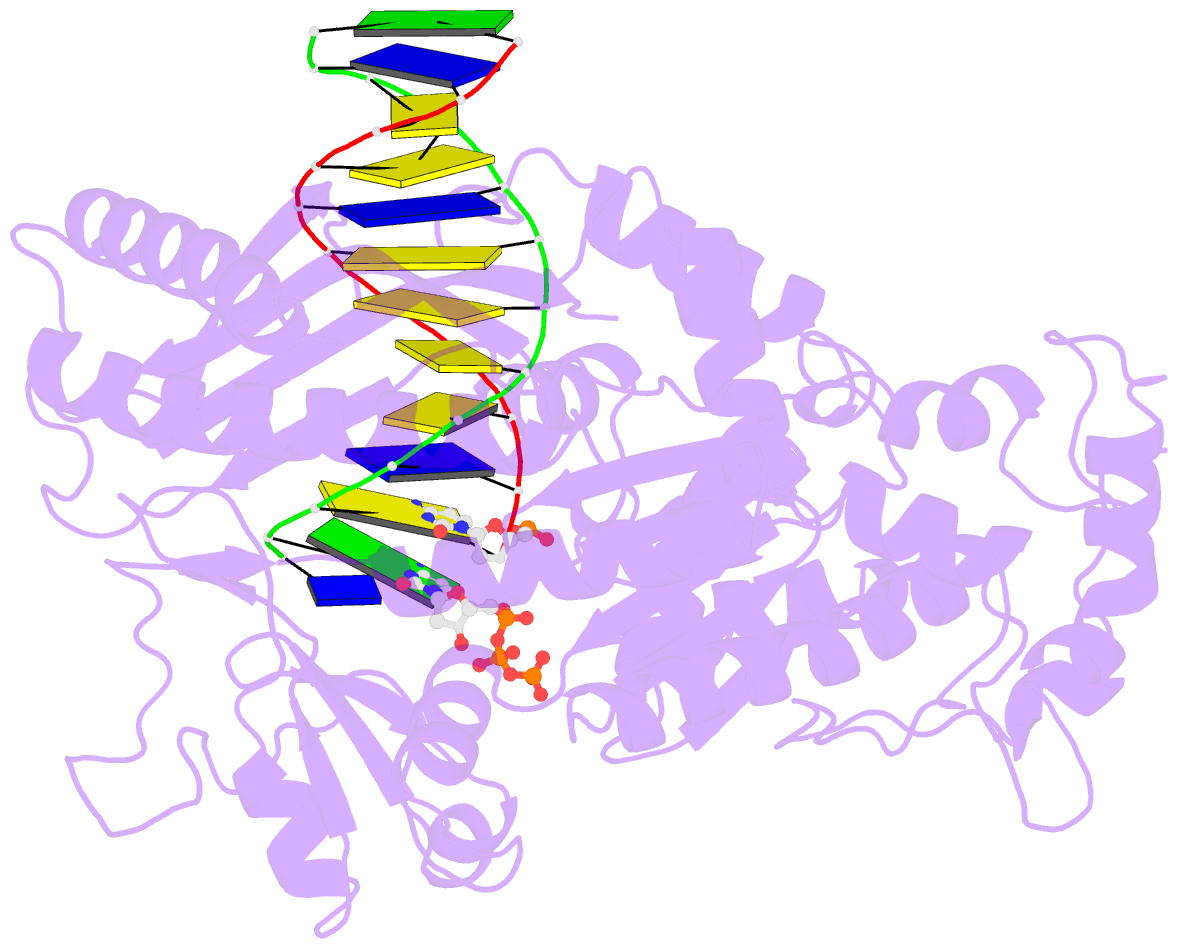
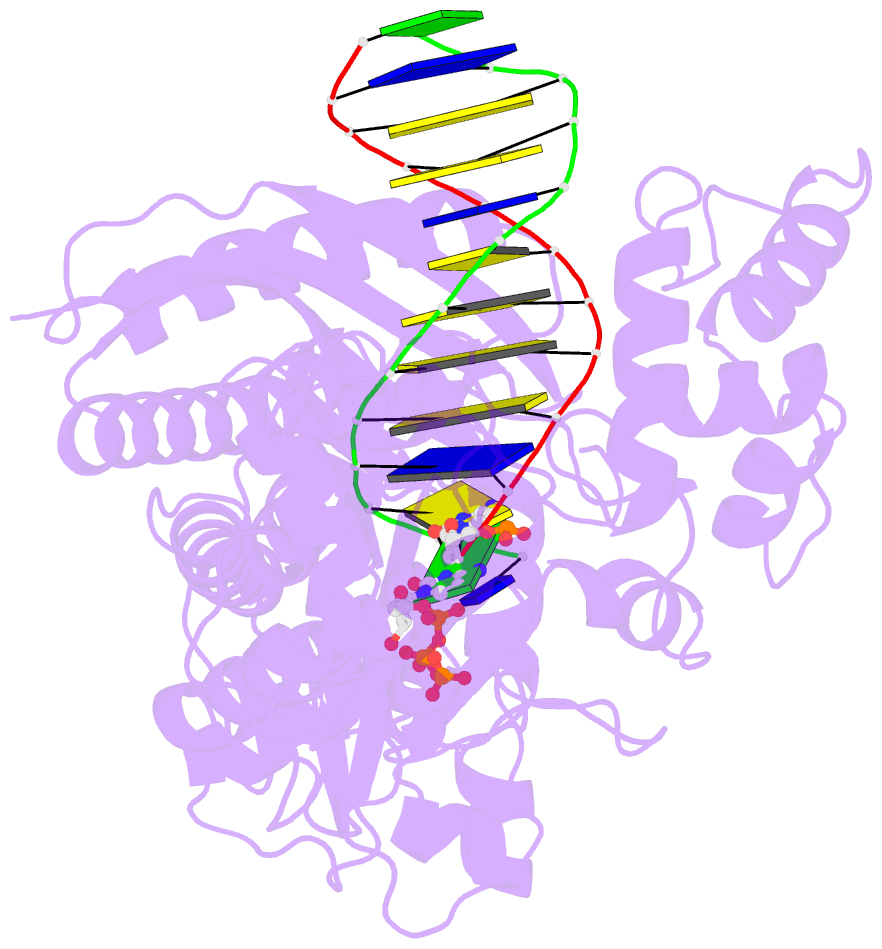
- Yeast DNA polymerase eta inserting dctp opposite an 8oxog lesion
- Reference
- Silverstein TD, Jain R, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2010): "Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta." Structure, 18, 1463-1470. doi: 10.1016/j.str.2010.08.019.
- Abstract
- 7,8-dihydro-8-oxoguanine (8-oxoG) adducts are formed frequently by the attack of oxygen-free radicals on DNA. They are among the most mutagenic lesions in cells because of their dual coding potential, where, in addition to normal base-pairing of 8-oxoG(anti) with dCTP, 8-oxoG in the syn conformation can base pair with dATP, causing G to T transversions. We provide here for the first time a structural basis for the error-free replication of 8-oxoG lesions by yeast DNA polymerase η (Polη). We show that the open active site cleft of Polη can accommodate an 8-oxoG lesion in the anti conformation with only minimal changes to the polymerase and the bound DNA: at both the insertion and post-insertion steps of lesion bypass. Importantly, the active site geometry remains the same as in the undamaged complex and provides a basis for the ability of Polη to prevent the mutagenic replication of 8-oxoG lesions in cells.