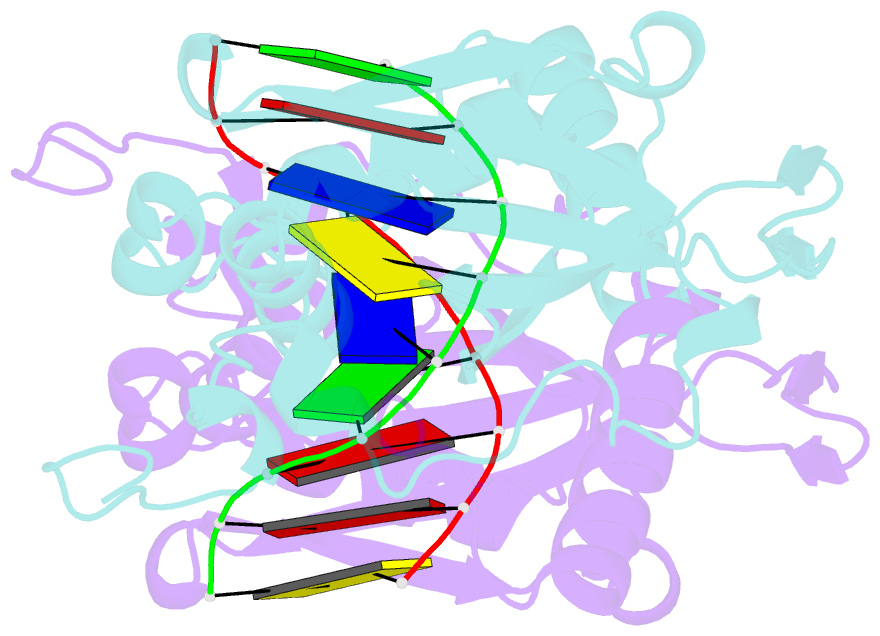
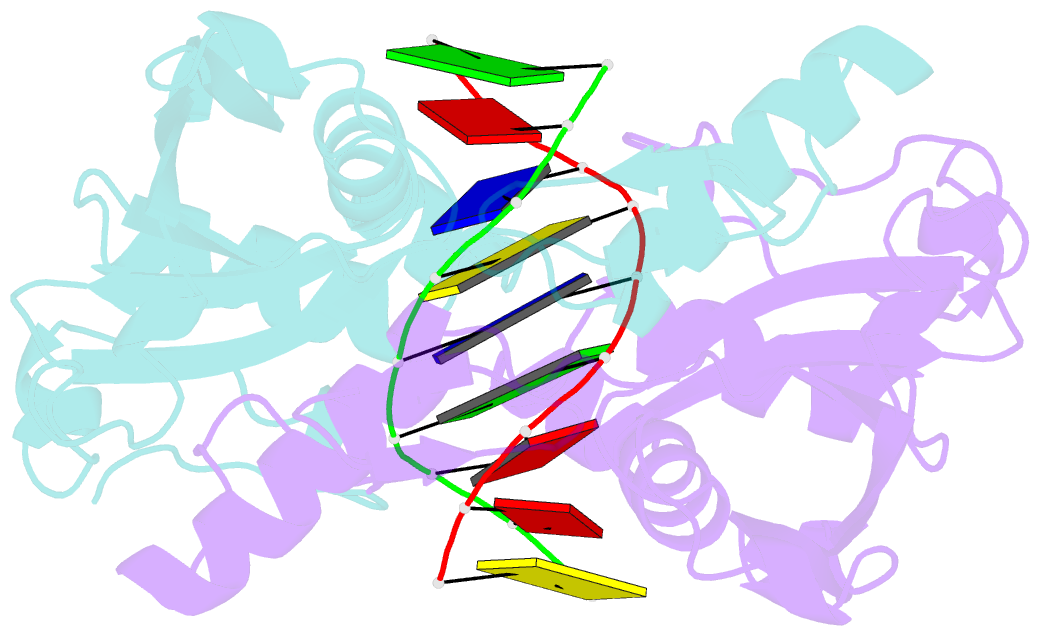
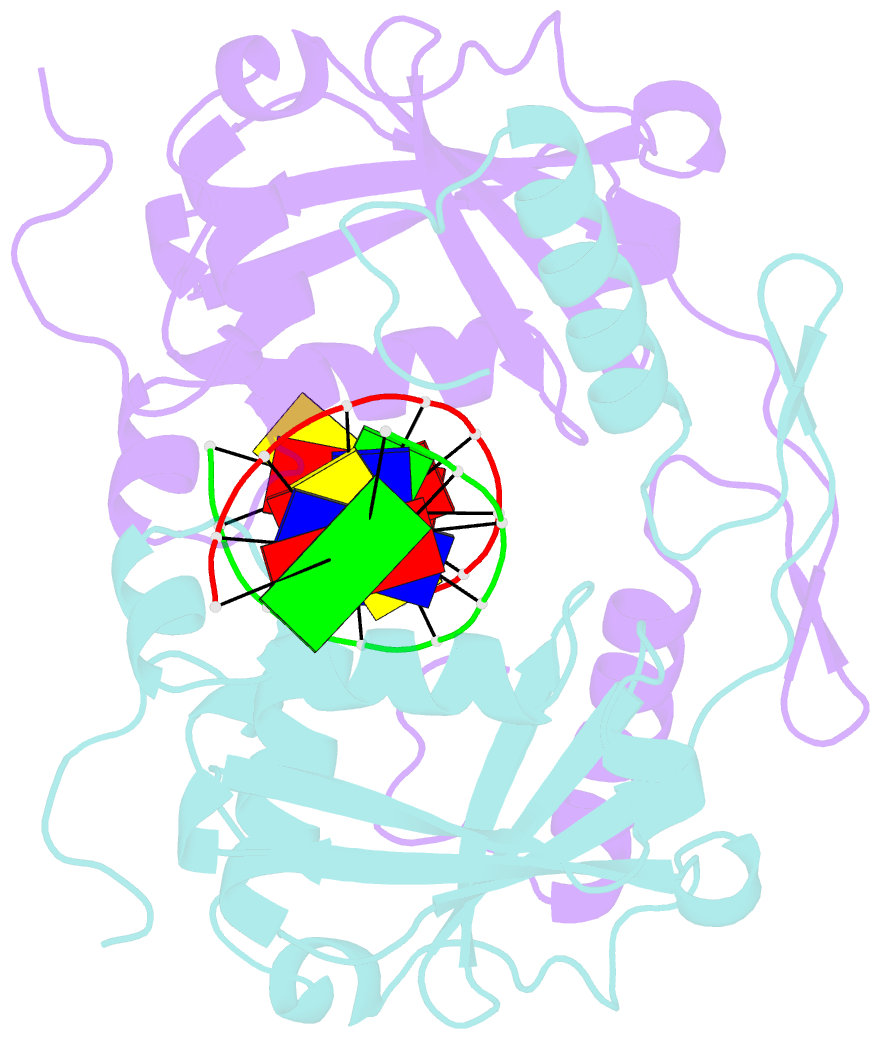
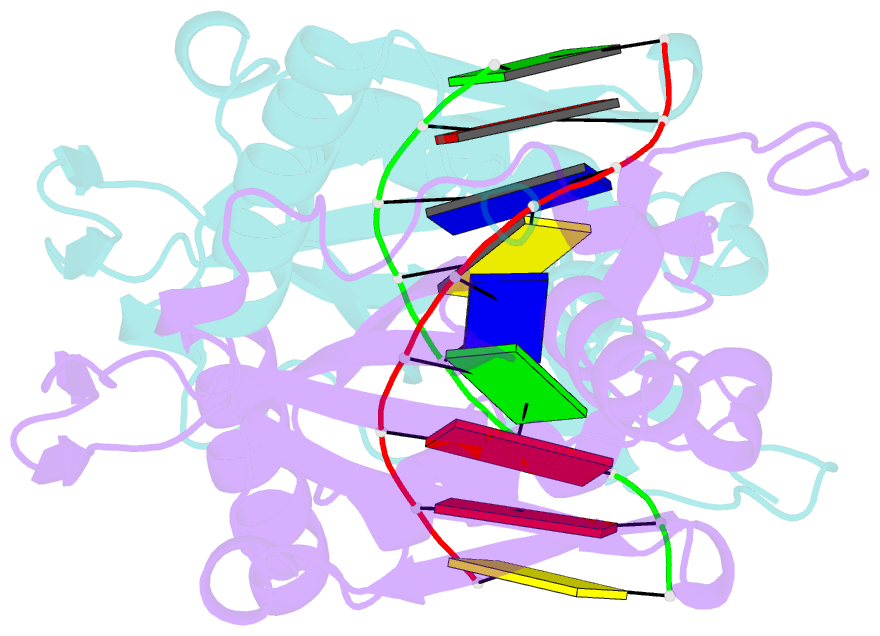
Summary information and primary citation
- PDB-id
- 3oqg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (1.75 Å)
- Summary
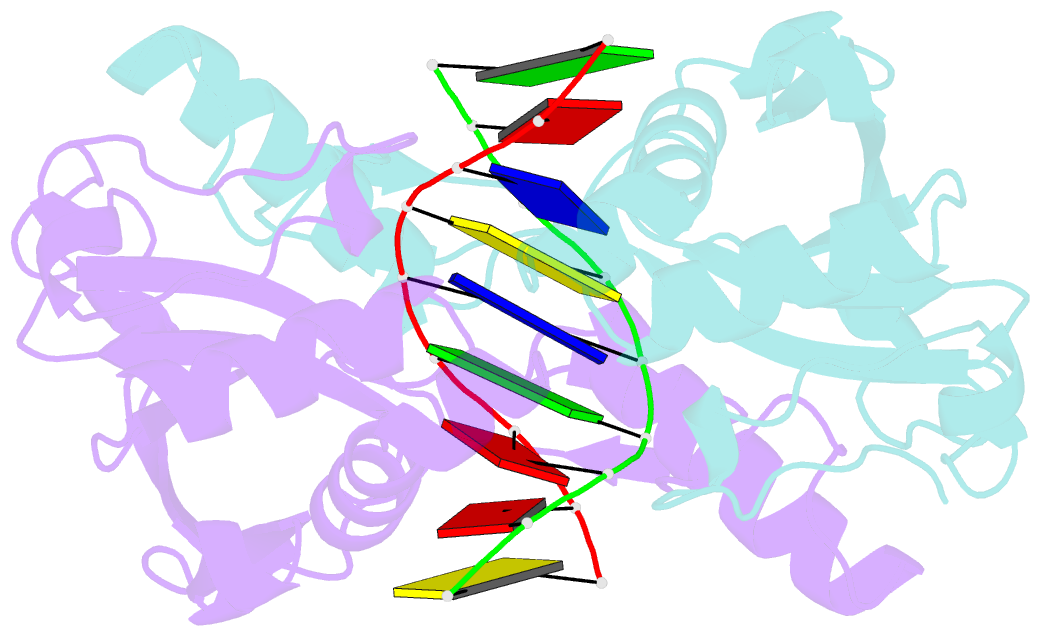
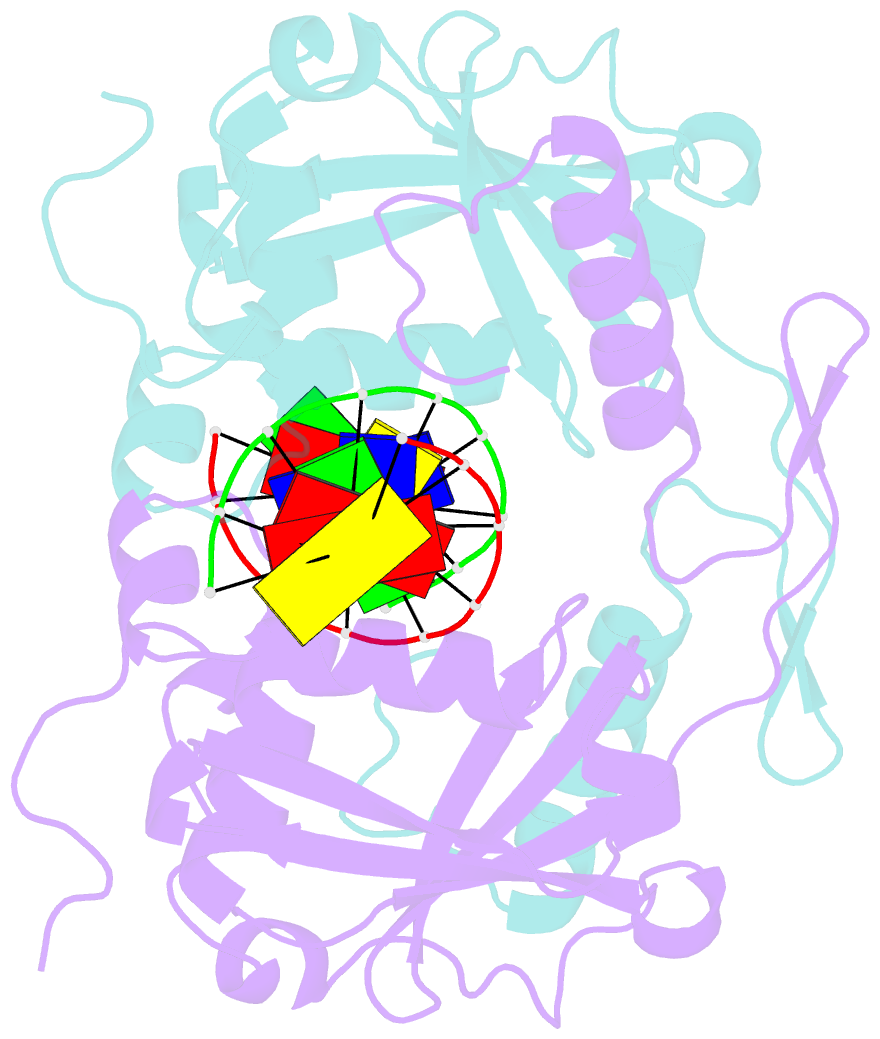
- Restriction endonuclease hpy188i in complex with substrate DNA
- Reference
- Sokolowska M, Czapinska H, Bochtler M (2011): "Hpy188I-DNA pre- and post-cleavage complexes--snapshots of the GIY-YIG nuclease mediated catalysis." Nucleic Acids Res., 39, 1554-1564. doi: 10.1093/nar/gkq821.
- Abstract
- The GIY-YIG nuclease domain is present in all kingdoms of life and has diverse functions. It is found in the eukaryotic flap endonuclease and Holliday junction resolvase Slx1-Slx4, the prokaryotic nucleotide excision repair proteins UvrC and Cho, and in proteins of 'selfish' genetic elements. Here we present the structures of the ternary pre- and post-cleavage complexes of the type II GIY-YIG restriction endonuclease Hpy188I with DNA and a surrogate or catalytic metal ion, respectively. Our structures suggest that GIY-YIG nucleases catalyze DNA hydrolysis by a single substitution reaction. They are consistent with a previous proposal that a tyrosine residue (which we expect to occur in its phenolate form) acts as a general base for the attacking water molecule. In contrast to the earlier proposal, our data identify the general base with the GIY and not the YIG tyrosine. A conserved glutamate residue (Glu149 provided in trans in Hpy188I) anchors a single metal cation in the active site. This metal ion contacts the phosphate proS oxygen atom and the leaving group 3'-oxygen atom, presumably to facilitate its departure. Taken together, our data reveal striking analogy in the absence of homology between GIY-YIG and ββα-Me nucleases.