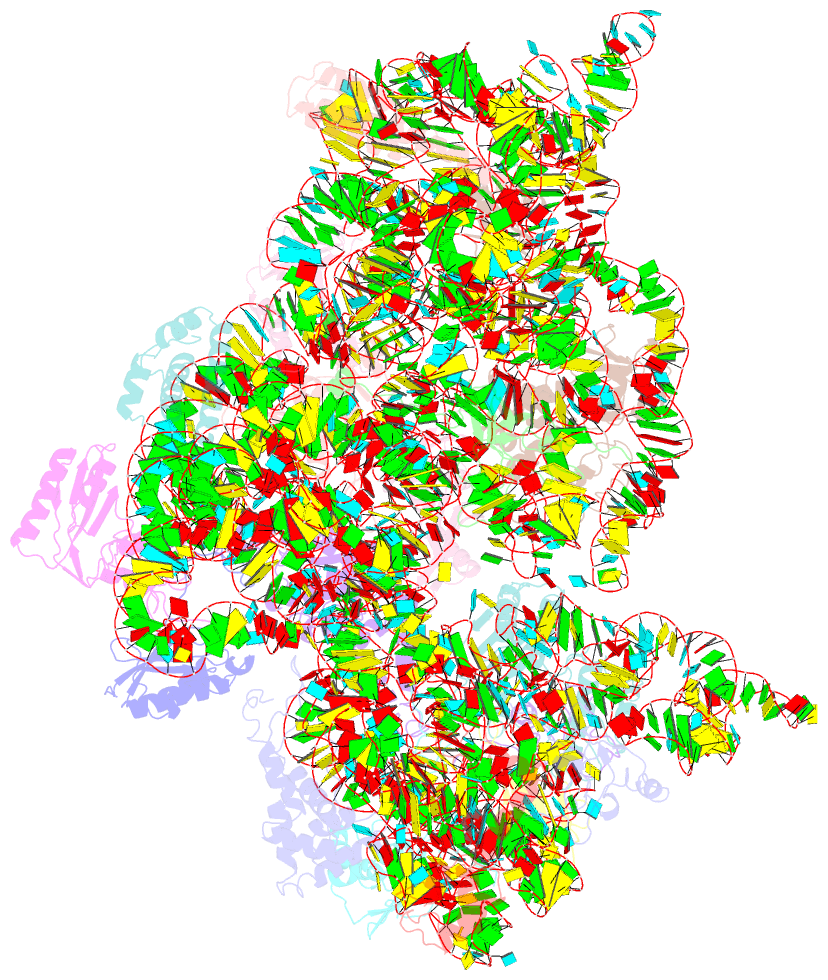
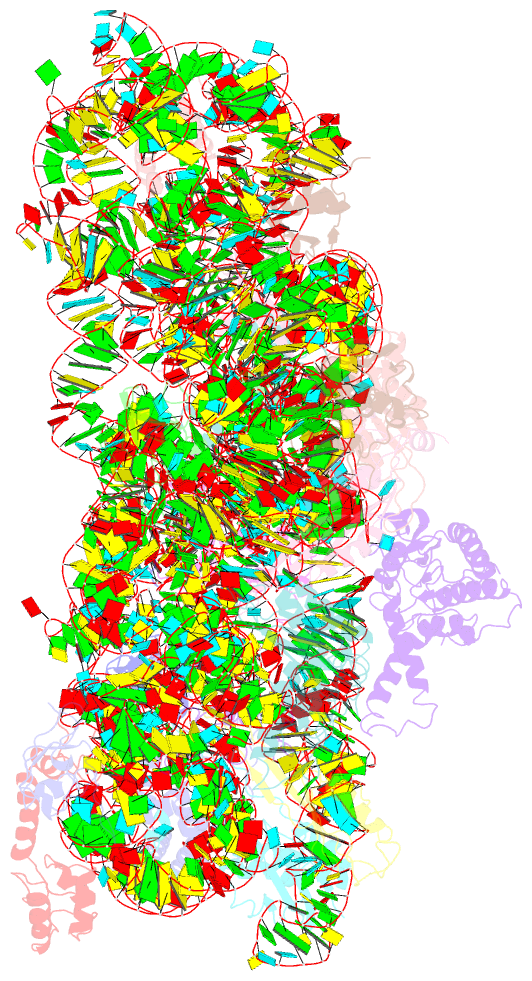
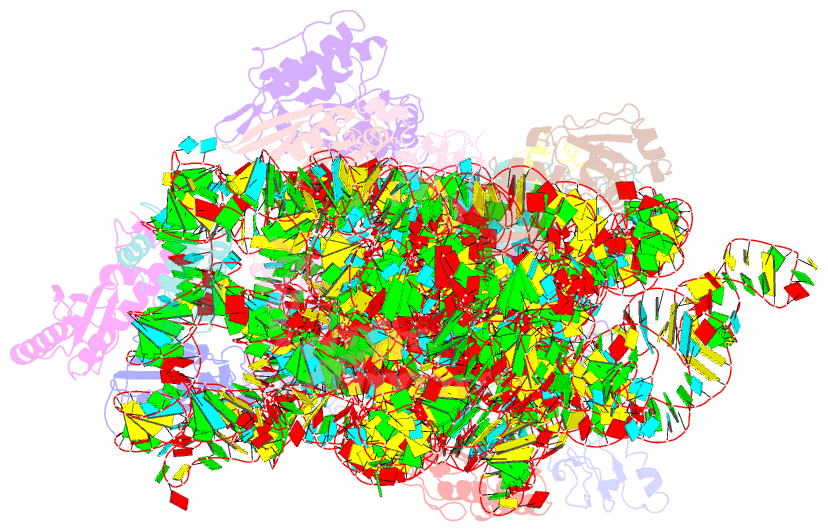
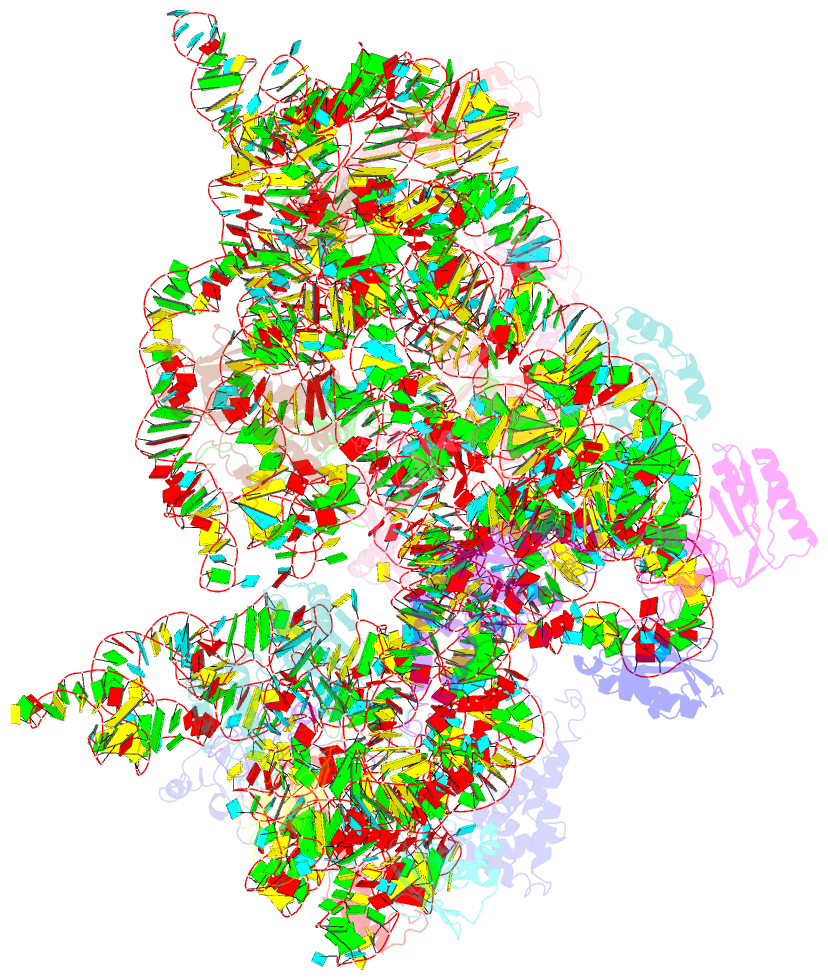
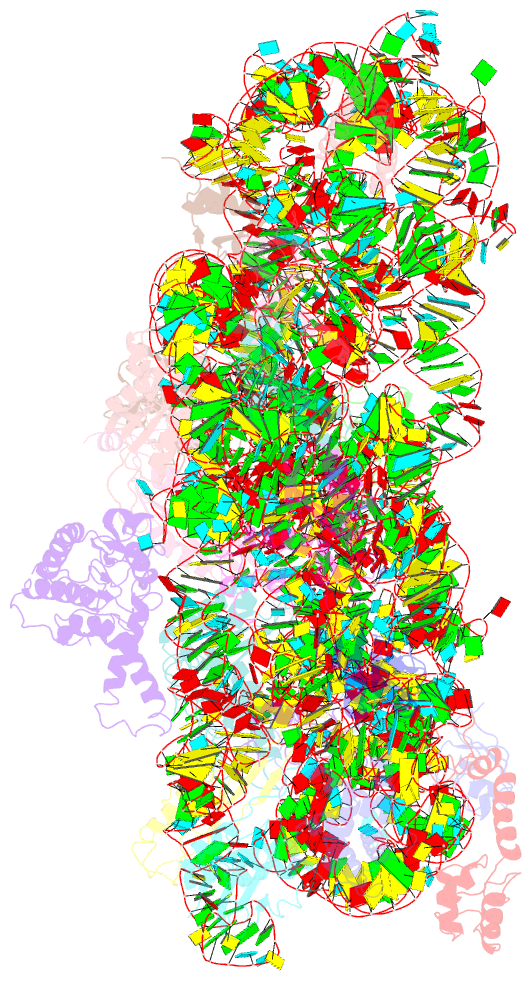
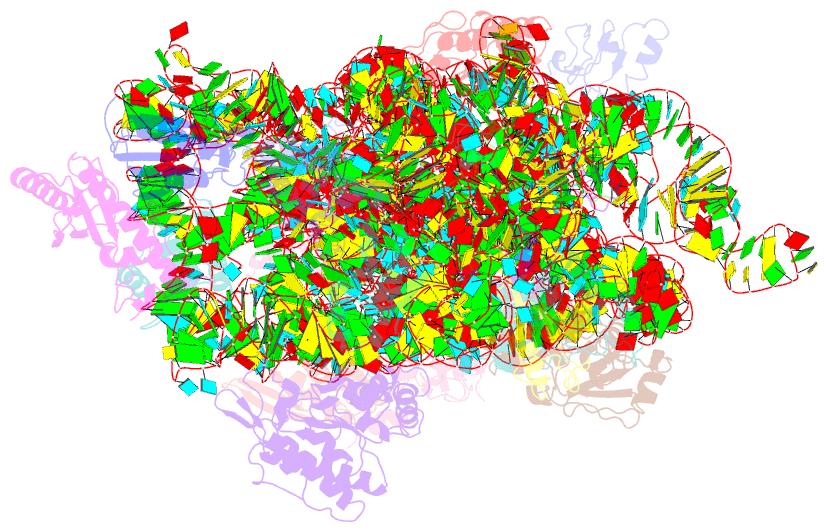
Summary information and primary citation
- PDB-id
- 3oto; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.69 Å)
- Summary
- Crystal structure of the 30s ribosomal subunit from a ksga mutant of thermus thermophilus (hb8)
- Reference
- Demirci H, Murphy F, Belardinelli R, Kelley AC, Ramakrishnan V, Gregory ST, Dahlberg AE, Jogl G (2010): "Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function." Rna, 16, 2319-2324. doi: 10.1261/rna.2357210.
- Abstract
- All organisms incorporate post-transcriptional modifications into ribosomal RNA, influencing ribosome assembly and function in ways that are poorly understood. The most highly conserved modification is the dimethylation of two adenosines near the 3' end of the small subunit rRNA. Lack of these methylations due to deficiency in the KsgA methyltransferase stimulates translational errors during both the initiation and elongation phases of protein synthesis and confers resistance to the antibiotic kasugamycin. Here, we present the X-ray crystal structure of the Thermus thermophilus 30S ribosomal subunit lacking these dimethylations. Our data indicate that the KsgA-directed methylations facilitate structural rearrangements in order to establish a functionally optimum subunit conformation during the final stages of ribosome assembly.