Summary information and primary citation
- PDB-id
- 3p49; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.55 Å)
- Summary
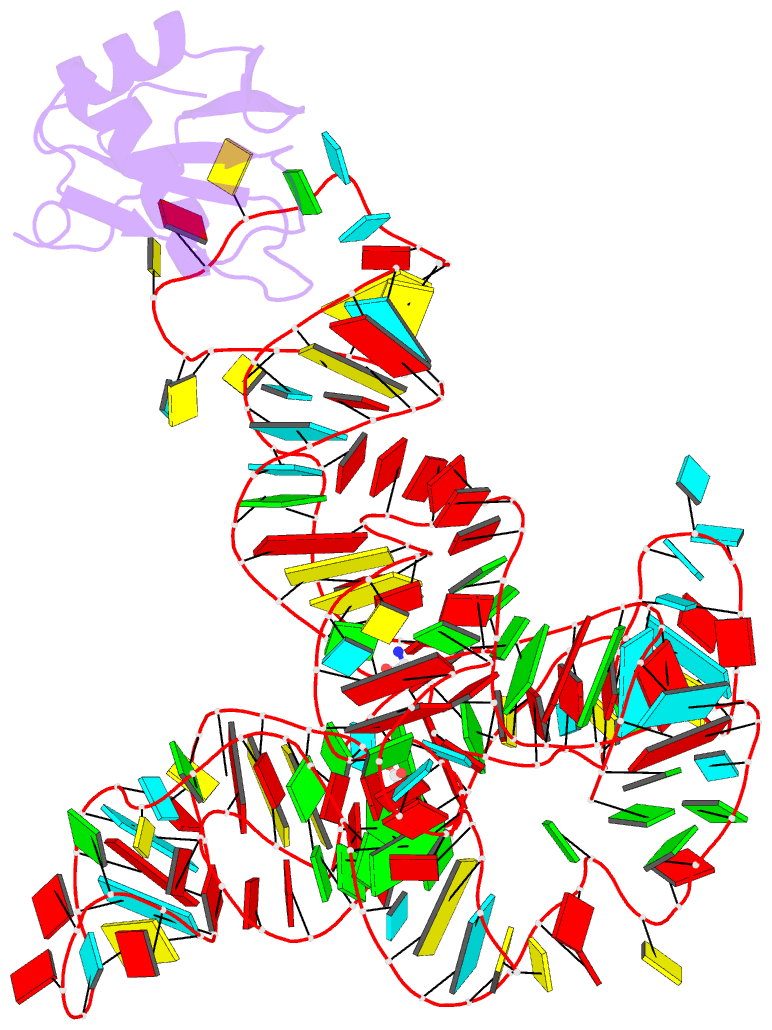
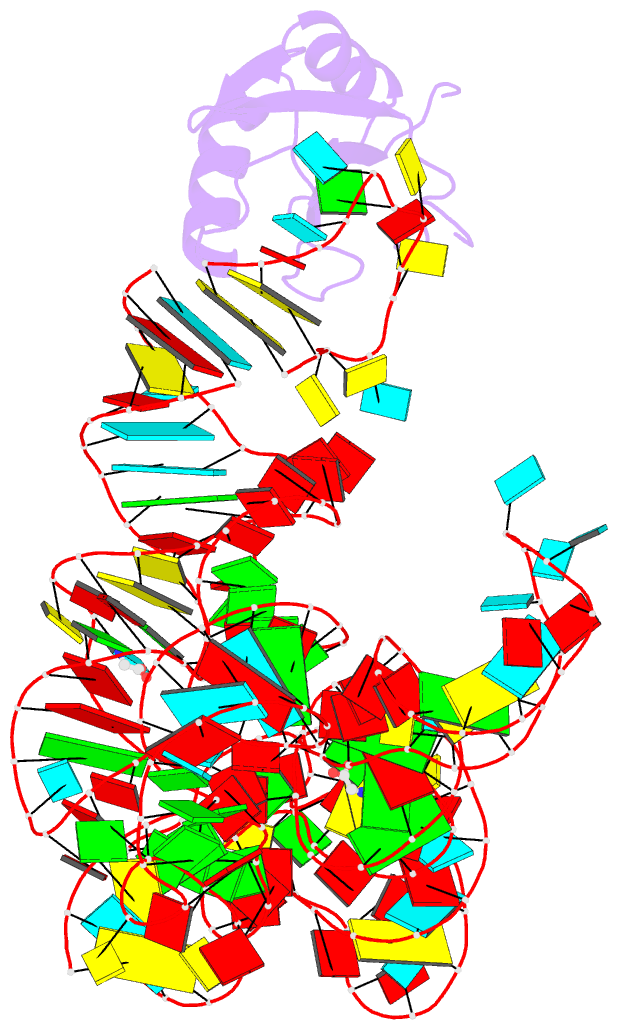
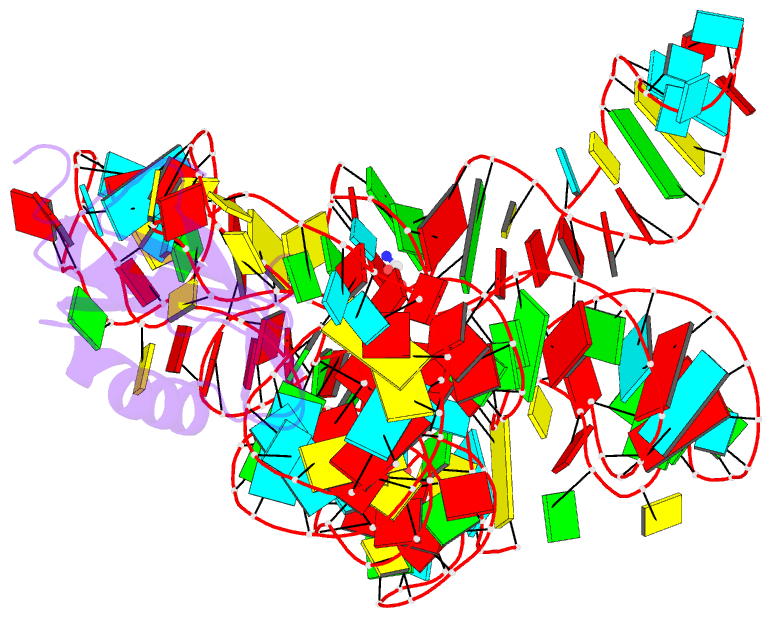
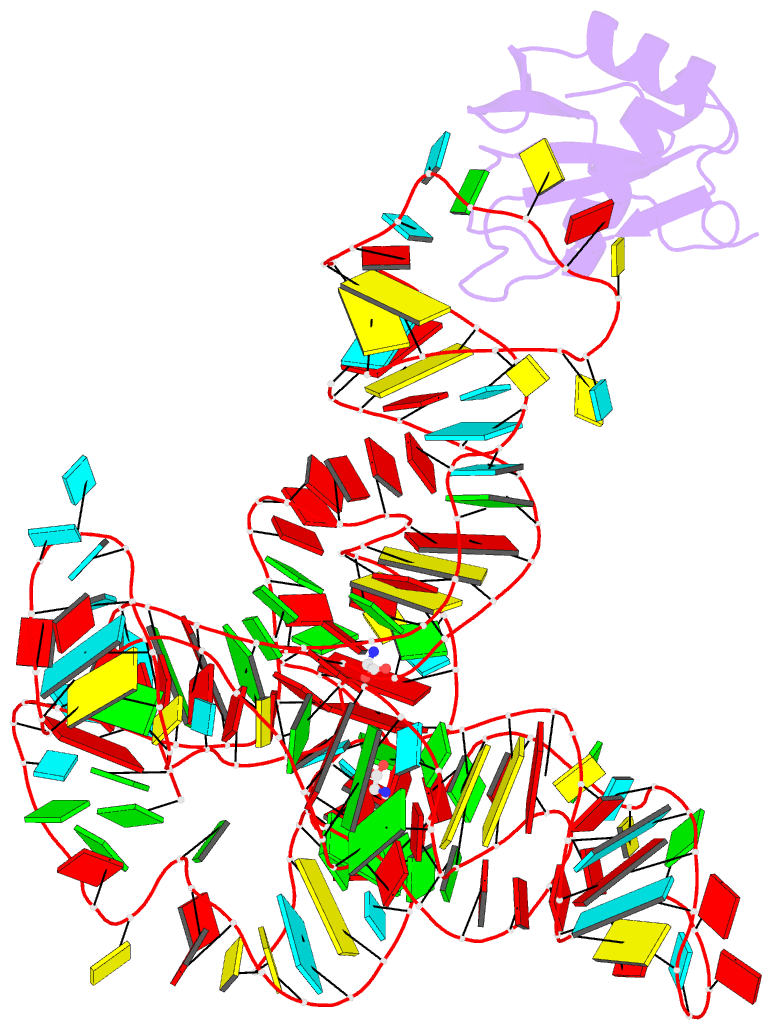
- Crystal structure of a glycine riboswitch from fusobacterium nucleatum
- Reference
- Butler EB, Xiong Y, Wang J, Strobel SA (2011): "Structural basis of cooperative ligand binding by the glycine riboswitch." Chem.Biol., 18, 293-298. doi: 10.1016/j.chembiol.2011.01.013.
- Abstract
- The glycine riboswitch regulates gene expression through the cooperative recognition of its amino acid ligand by a tandem pair of aptamers. A 3.6 Å crystal structure of the tandem riboswitch from the glycine permease operon of Fusobacterium nucleatum reveals the glycine binding sites and an extensive network of interactions, largely mediated by asymmetric A-minor contacts, that serve to communicate ligand binding status between the aptamers. These interactions provide a structural basis for how the glycine riboswitch cooperatively regulates gene expression.