Summary information and primary citation
- PDB-id
- 3po3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA
- Method
- X-ray (3.3 Å)
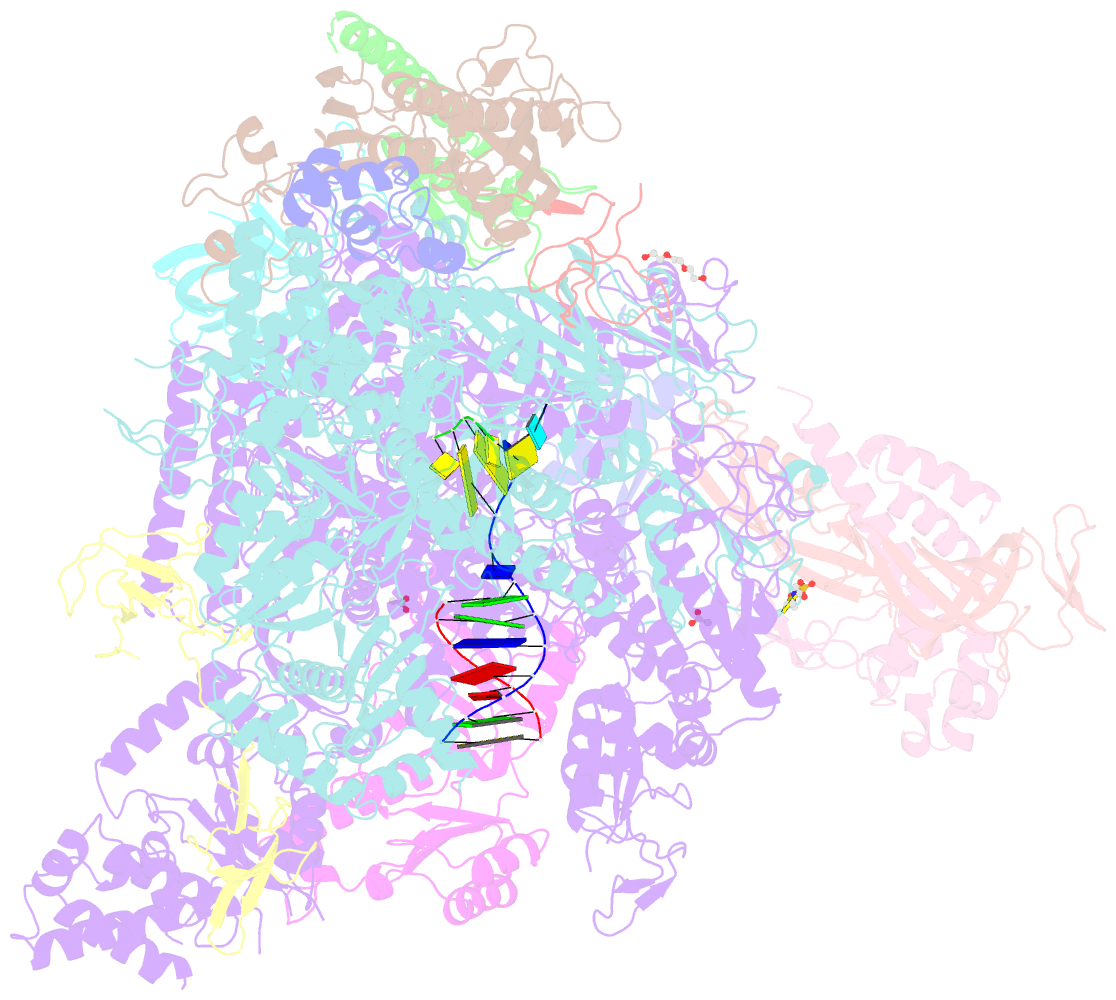
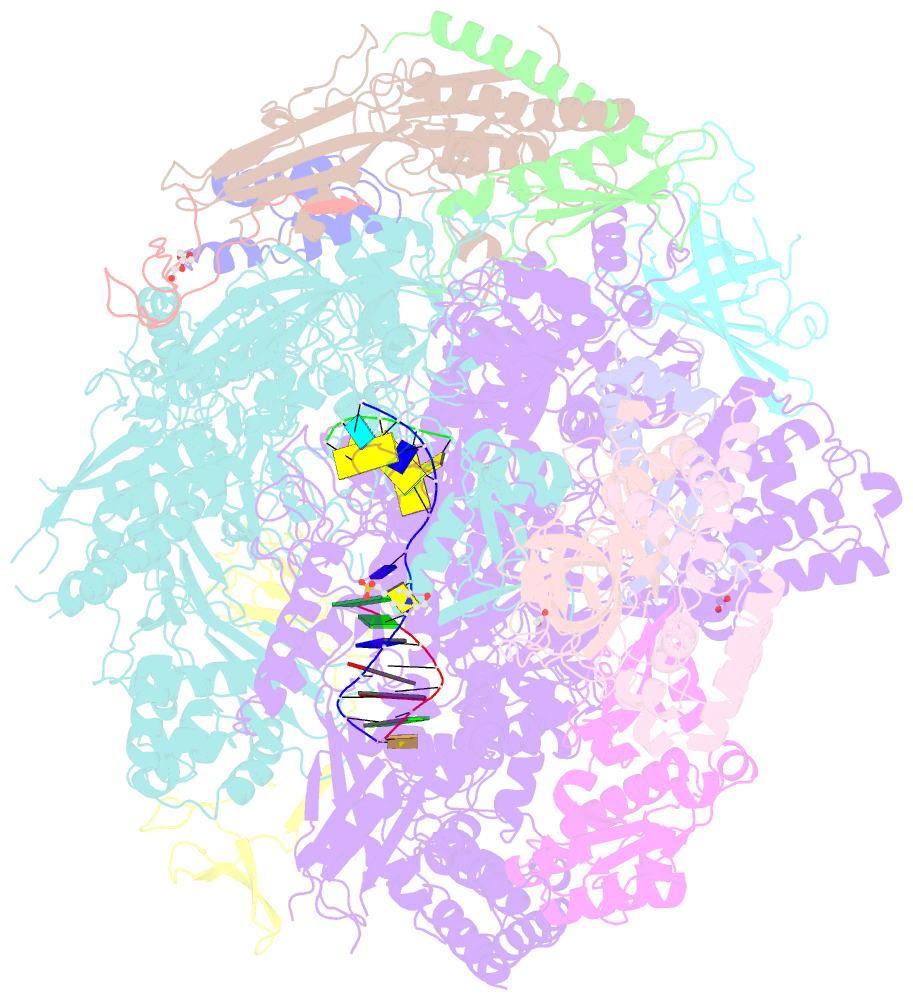
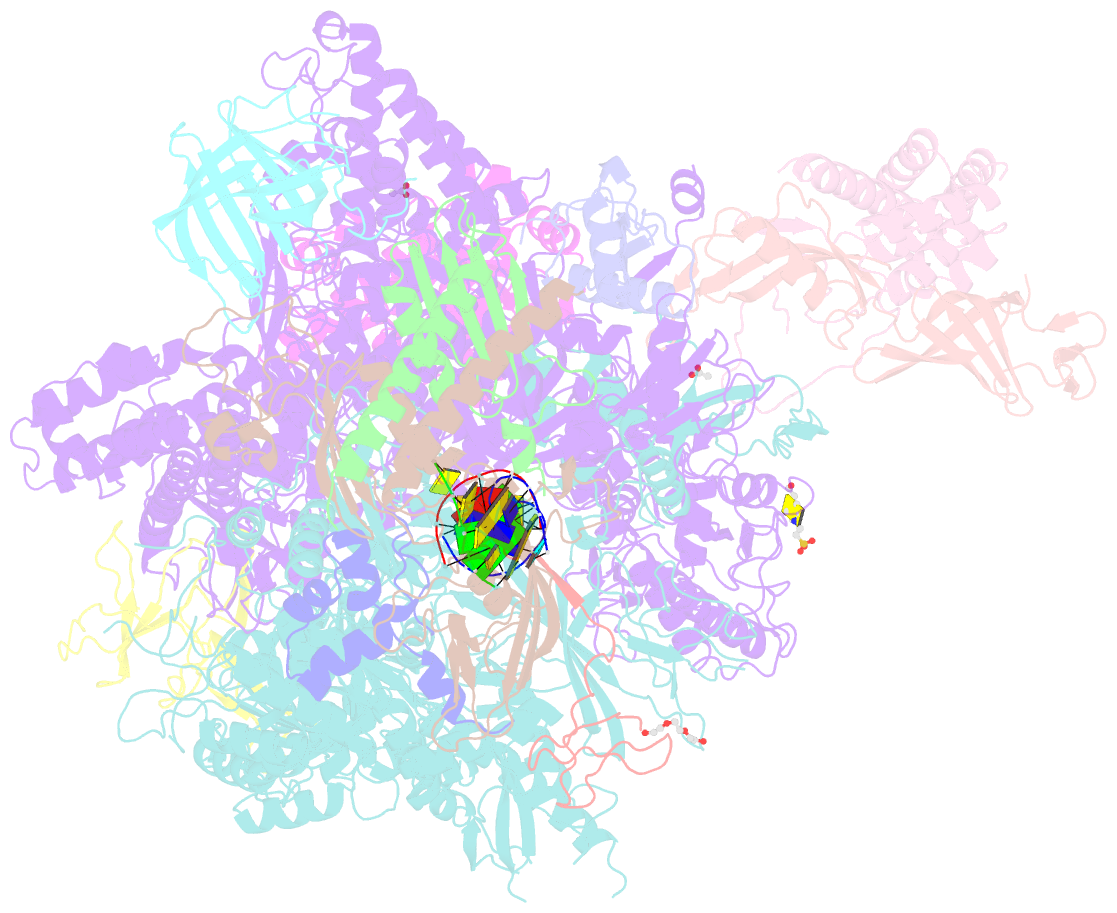
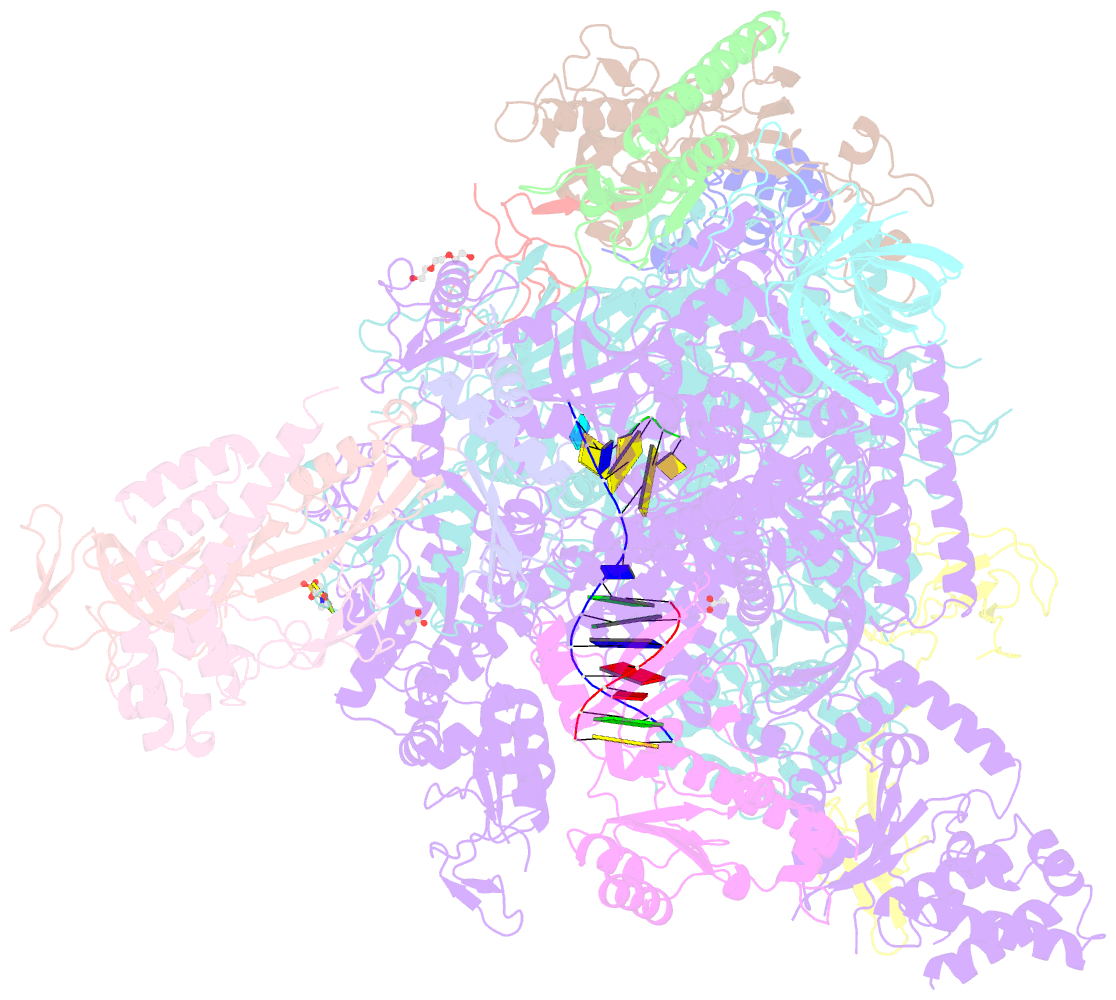
- Summary
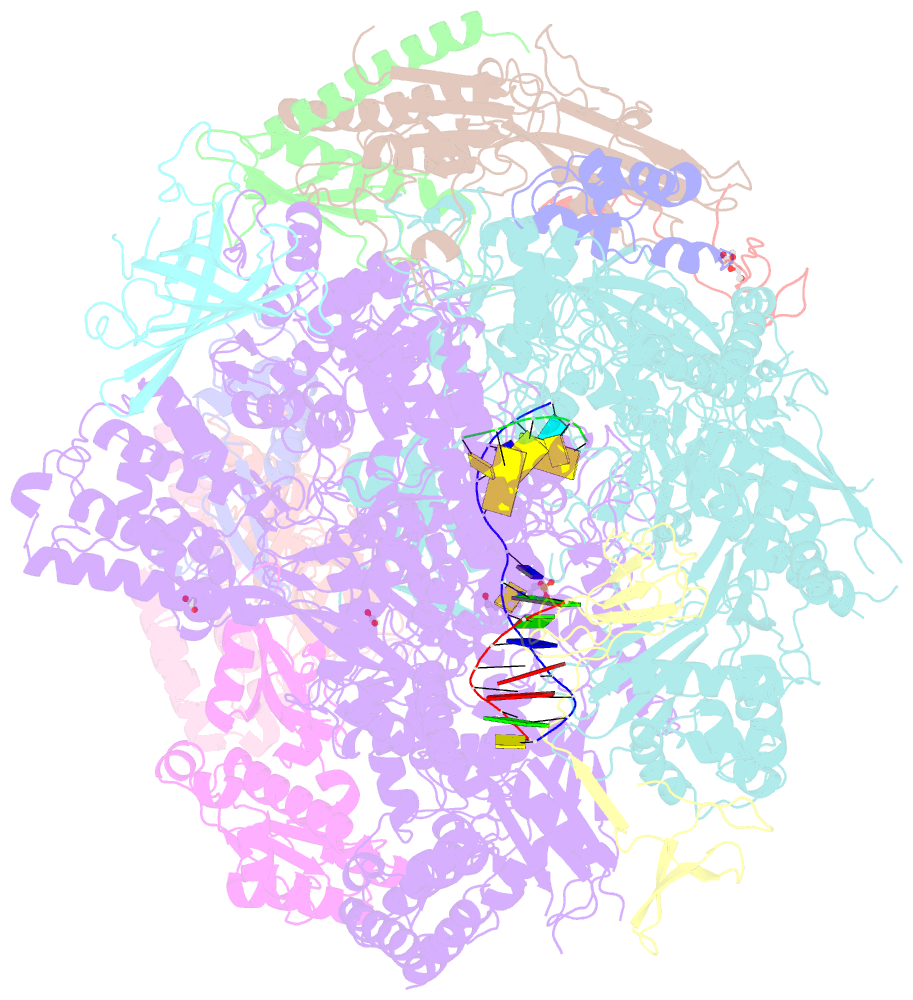
- Arrested RNA polymerase ii reactivation intermediate
- Reference
- Cheung AC, Cramer P (2011): "Structural basis of RNA polymerase II backtracking, arrest and reactivation." Nature, 471, 249-253. doi: 10.1038/nature09785.
- Abstract
- During gene transcription, RNA polymerase (Pol) II moves forwards along DNA and synthesizes messenger RNA. However, at certain DNA sequences, Pol II moves backwards, and such backtracking can arrest transcription. Arrested Pol II is reactivated by transcription factor IIS (TFIIS), which induces RNA cleavage that is required for cell viability. Pol II arrest and reactivation are involved in transcription through nucleosomes and in promoter-proximal gene regulation. Here we present X-ray structures at 3.3 Å resolution of an arrested Saccharomyces cerevisiae Pol II complex with DNA and RNA, and of a reactivation intermediate that additionally contains TFIIS. In the arrested complex, eight nucleotides of backtracked RNA bind a conserved 'backtrack site' in the Pol II pore and funnel, trapping the active centre trigger loop and inhibiting mRNA elongation. In the reactivation intermediate, TFIIS locks the trigger loop away from backtracked RNA, displaces RNA from the backtrack site, and complements the polymerase active site with a basic and two acidic residues that may catalyse proton transfers during RNA cleavage. The active site is demarcated from the backtrack site by a 'gating tyrosine' residue that probably delimits backtracking. These results establish the structural basis of Pol II backtracking, arrest and reactivation, and provide a framework for analysing gene regulation during transcription elongation.