Summary information and primary citation
- PDB-id
- 3px6; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.59 Å)
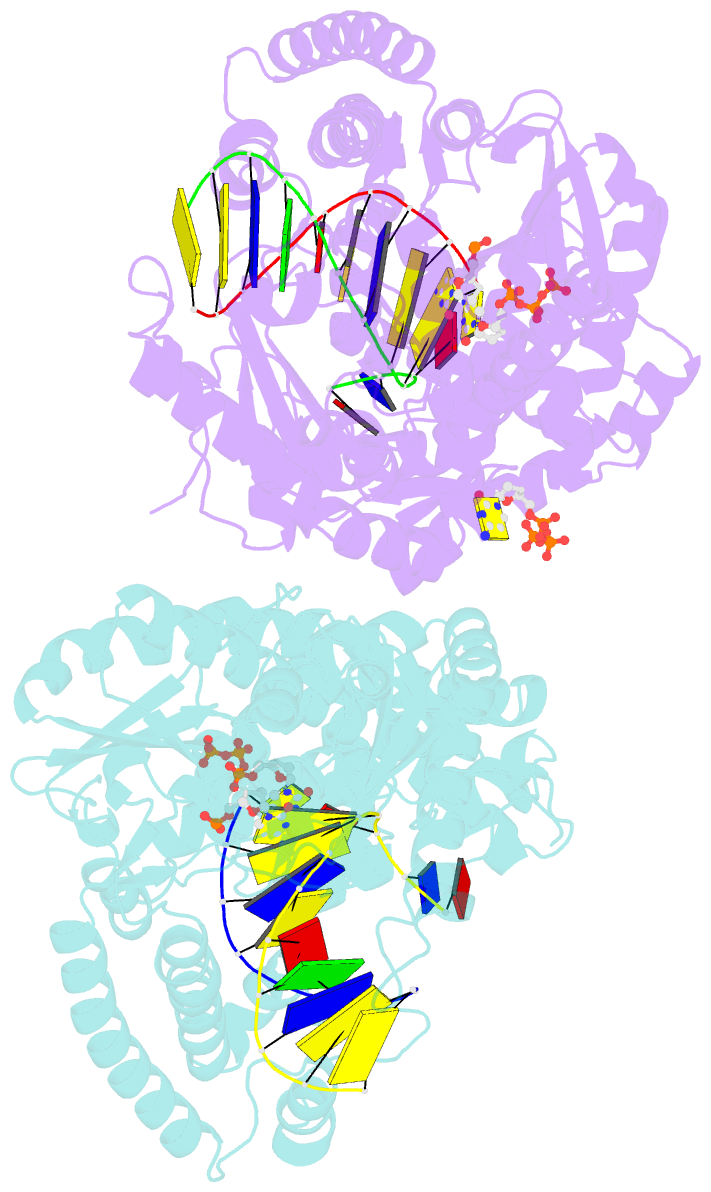
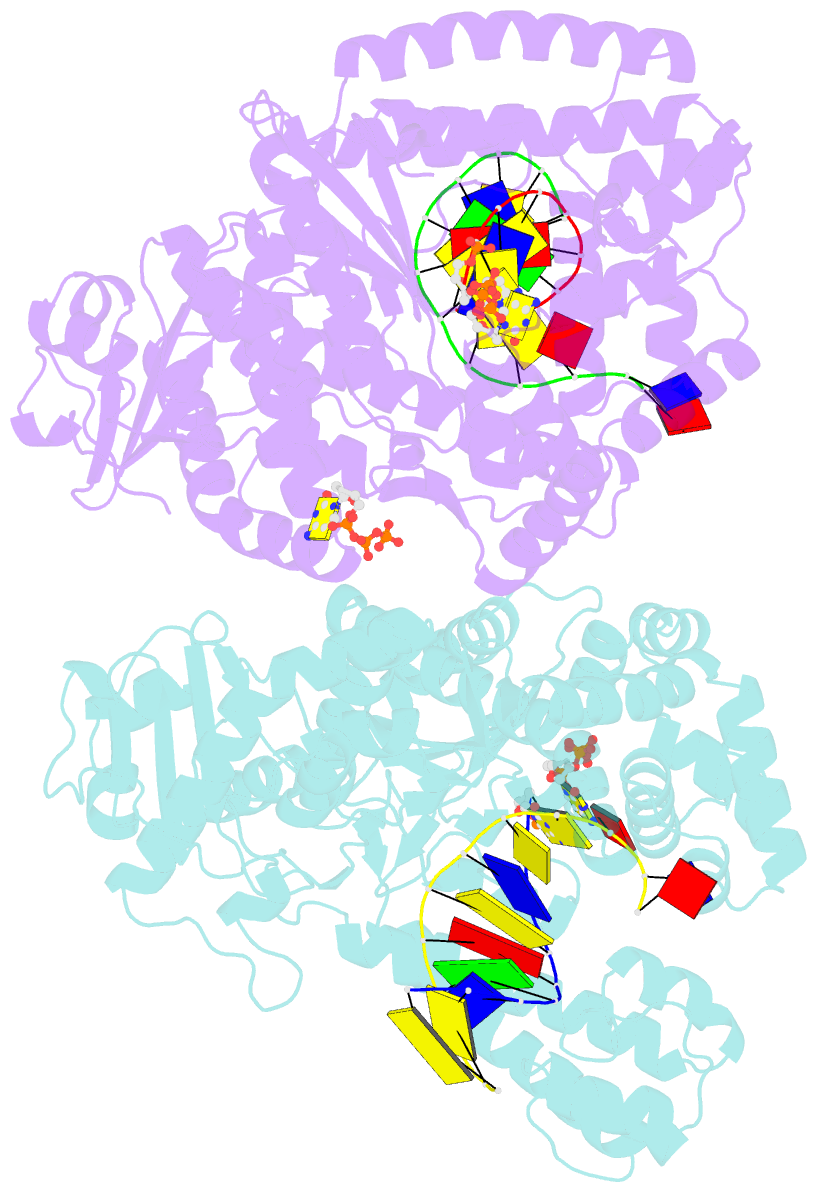
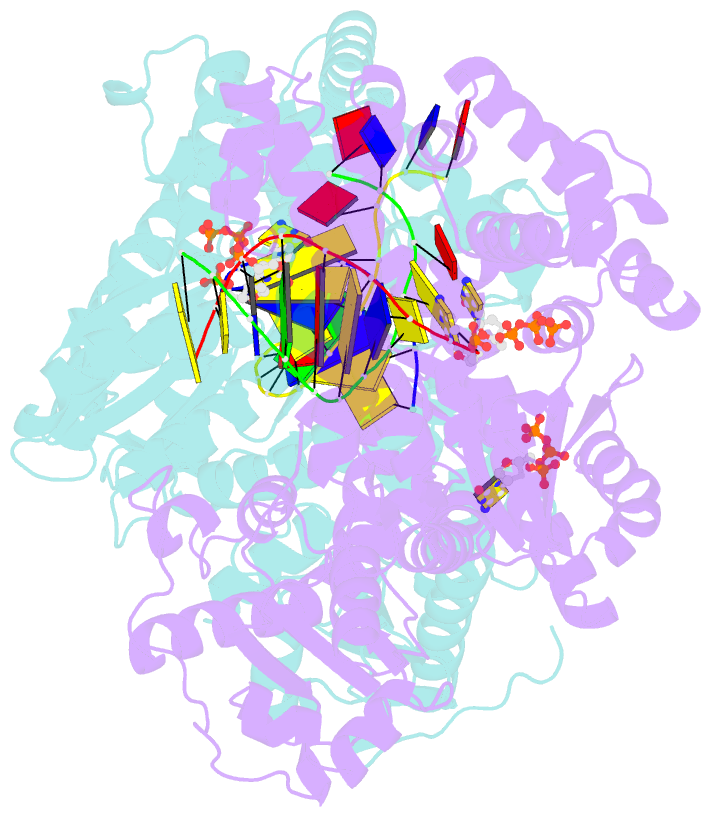
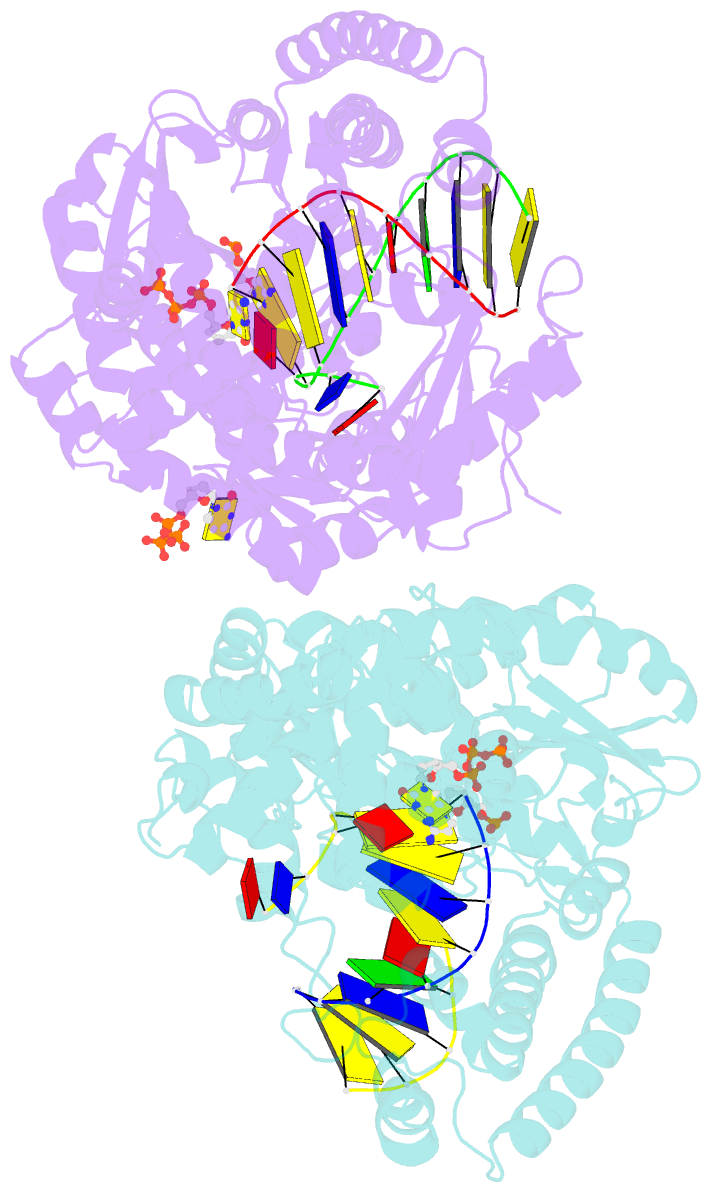
- Summary
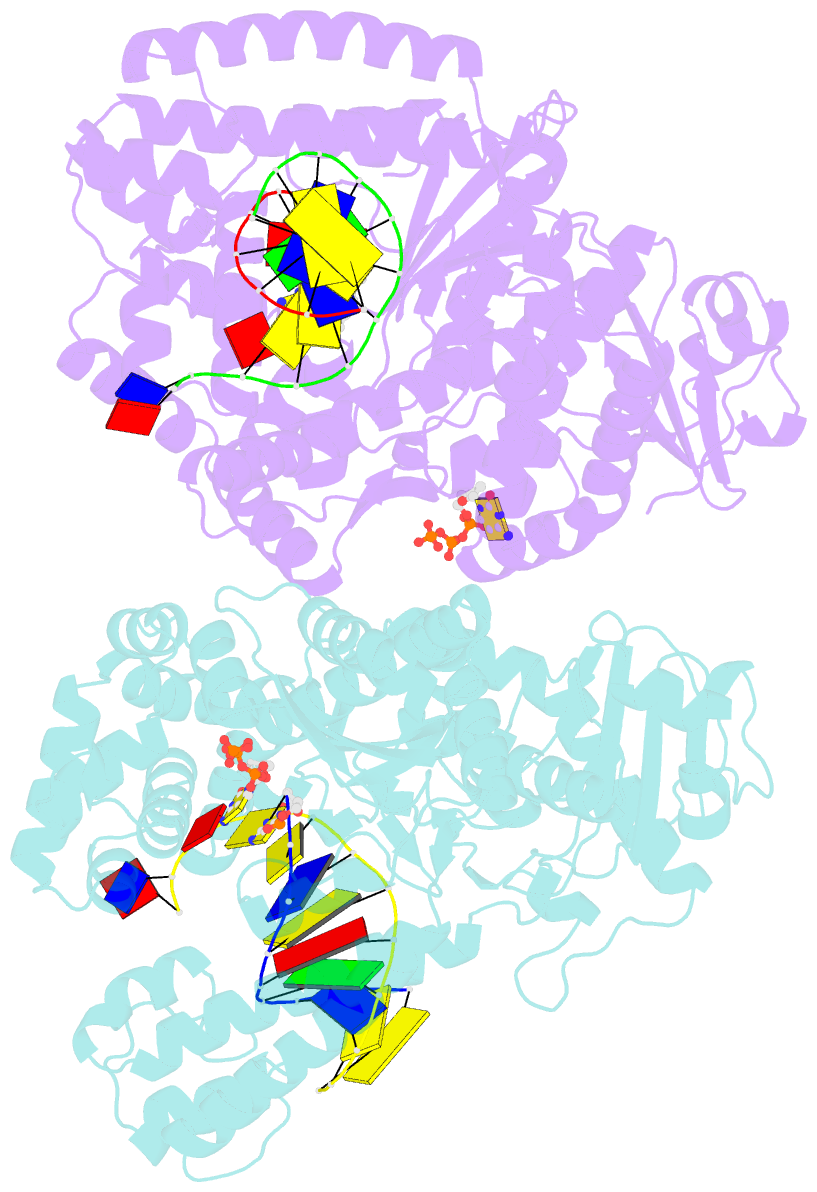
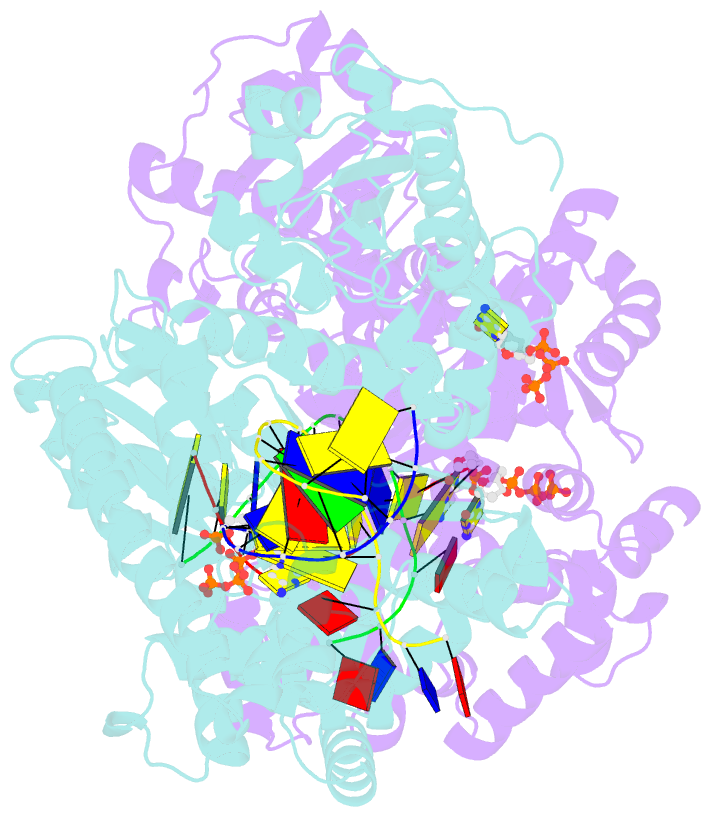
- Crystal structure of bacillus DNA polymerase i large fragment bound to DNA and ddctp-da mismatch (tautomer) in closed conformation
- Reference
- Wang W, Hellinga HW, Beese LS (2011): "Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis." Proc.Natl.Acad.Sci.USA, 108, 17644-17648. doi: 10.1073/pnas.1114496108.
- Abstract
- Even though high-fidelity polymerases copy DNA with remarkable accuracy, some base-pair mismatches are incorporated at low frequency, leading to spontaneous mutagenesis. Using high-resolution X-ray crystallographic analysis of a DNA polymerase that catalyzes replication in crystals, we observe that a C • A mismatch can mimic the shape of cognate base pairs at the site of incorporation. This shape mimicry enables the mismatch to evade the error detection mechanisms of the polymerase, which would normally either prevent mismatch incorporation or promote its nucleolytic excision. Movement of a single proton on one of the mismatched bases alters the hydrogen-bonding pattern such that a base pair forms with an overall shape that is virtually indistinguishable from a canonical, Watson-Crick base pair in double-stranded DNA. These observations provide structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis, a long-standing concept that has been difficult to demonstrate directly.