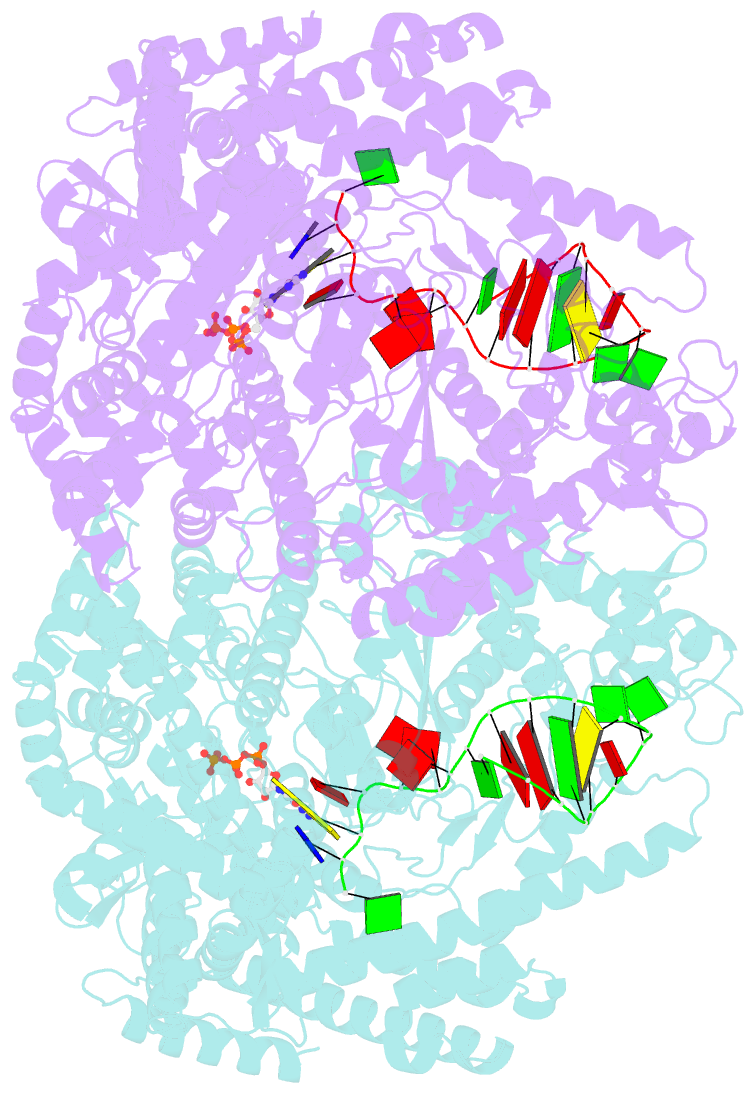
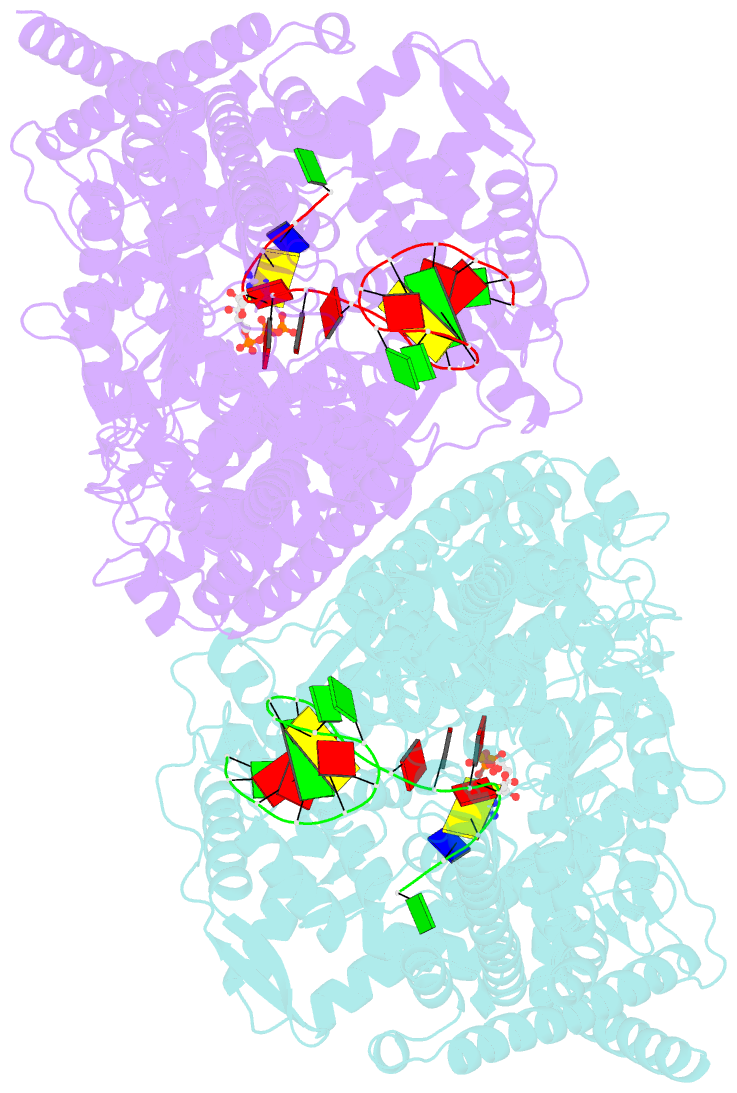
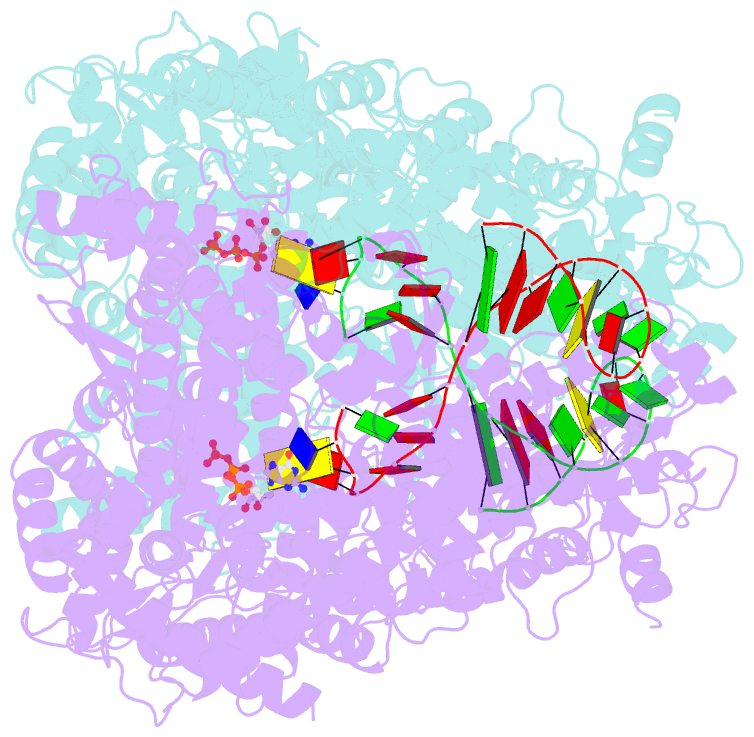
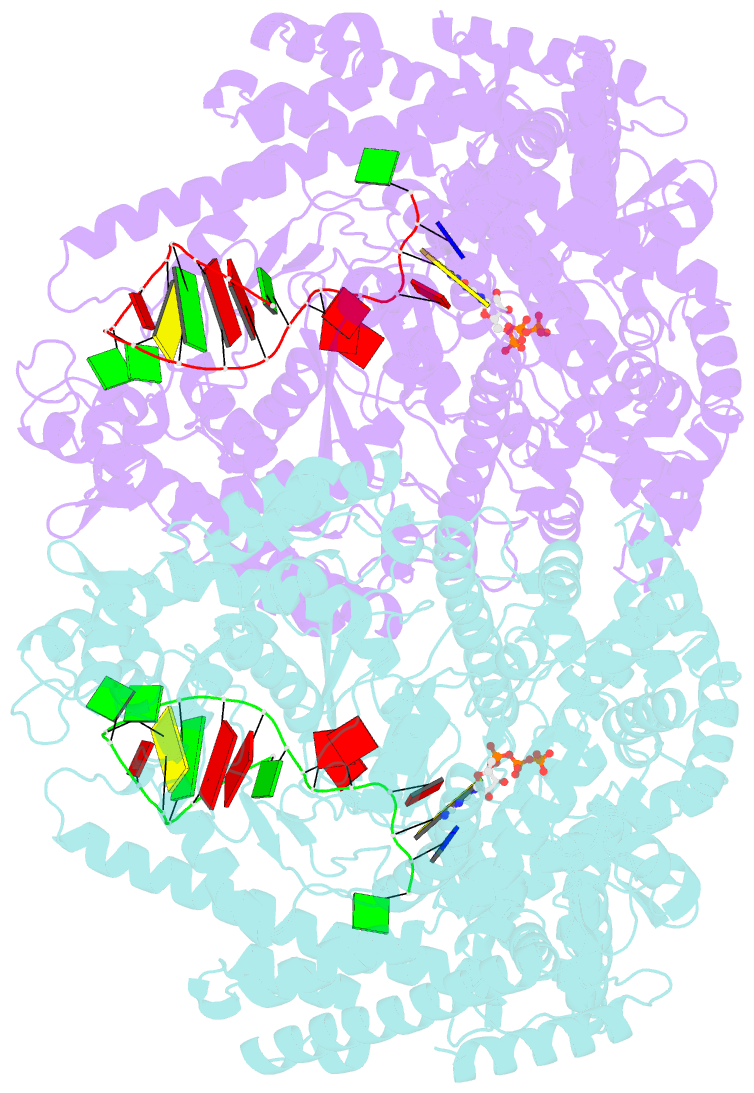
Summary information and primary citation
- PDB-id
- 3q0a; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.69 Å)
- Summary
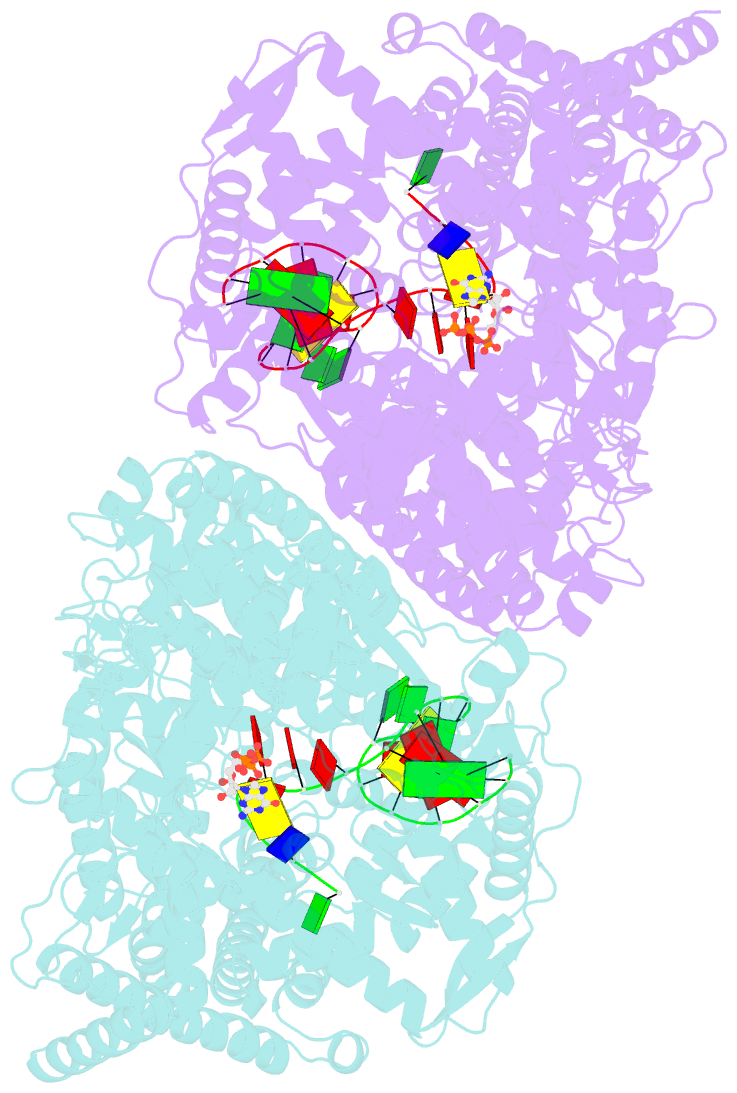
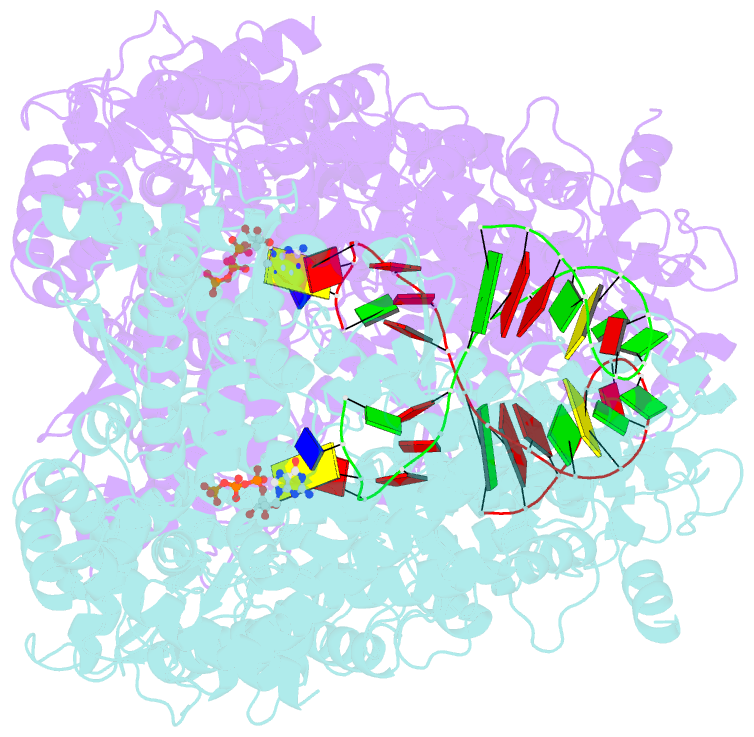
- X-ray crystal structure of the transcription initiation complex of the n4 mini-vrnap with p2 promoter: mismatch complex
- Reference
- Gleghorn ML, Davydova EK, Basu R, Rothman-Denes LB, Murakami KS (2011): "X-ray crystal structures elucidate the nucleotidyl transfer reaction of transcript initiation using two nucleotides." Proc.Natl.Acad.Sci.USA, 108, 3566-3571. doi: 10.1073/pnas.1016691108.
- Abstract
- We have determined the X-ray crystal structures of the pre- and postcatalytic forms of the initiation complex of bacteriophage N4 RNA polymerase that provide the complete set of atomic images depicting the process of transcript initiation by a single-subunit RNA polymerase. As observed during T7 RNA polymerase transcript elongation, substrate loading for the initiation process also drives a conformational change of the O-helix, but only the correct base pairing between the +2 substrate and DNA base is able to complete the O-helix conformational transition. Substrate binding also facilitates catalytic metal binding that leads to alignment of the reactive groups of substrates for the nucleotidyl transfer reaction. Although all nucleic acid polymerases use two divalent metals for catalysis, they differ in the requirements and the timing of binding of each metal. In the case of bacteriophage RNA polymerase, we propose that catalytic metal binding is the last step before the nucleotidyl transfer reaction.