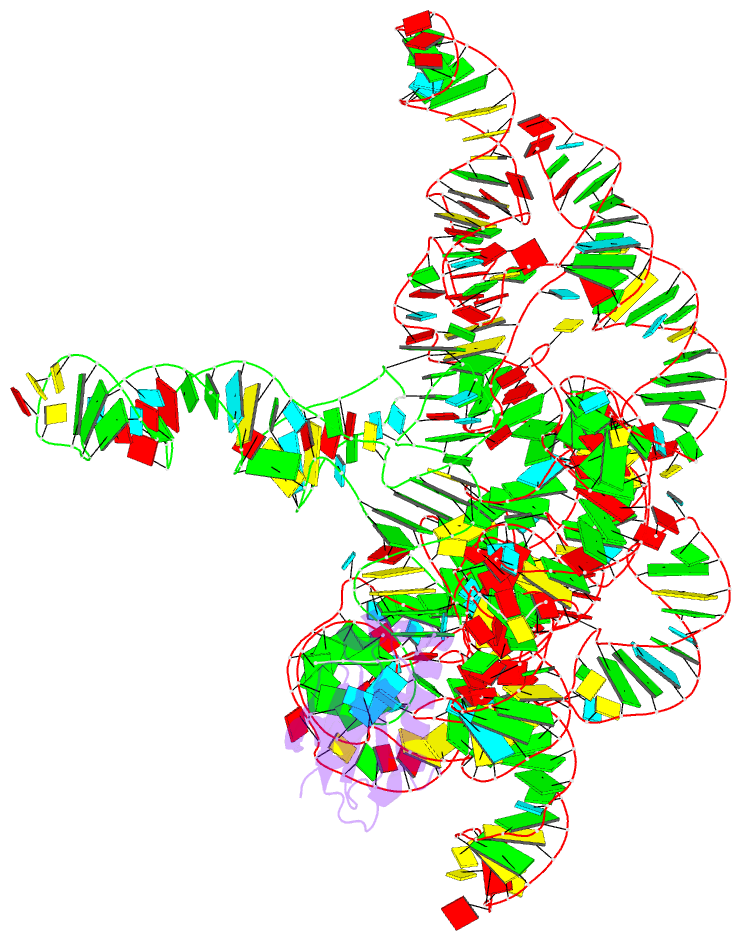
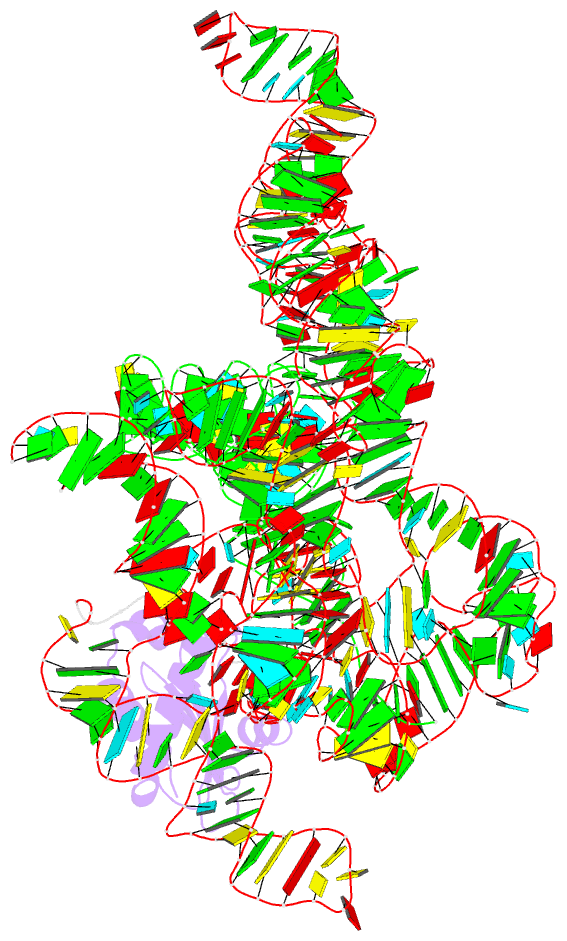
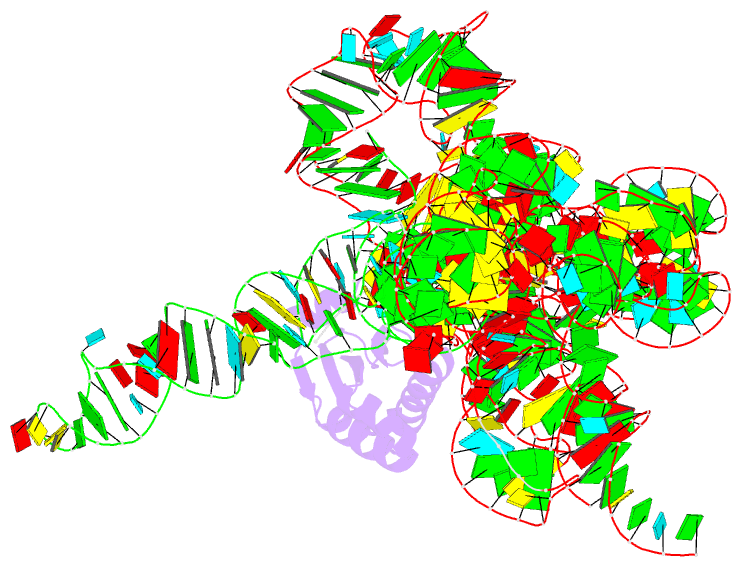
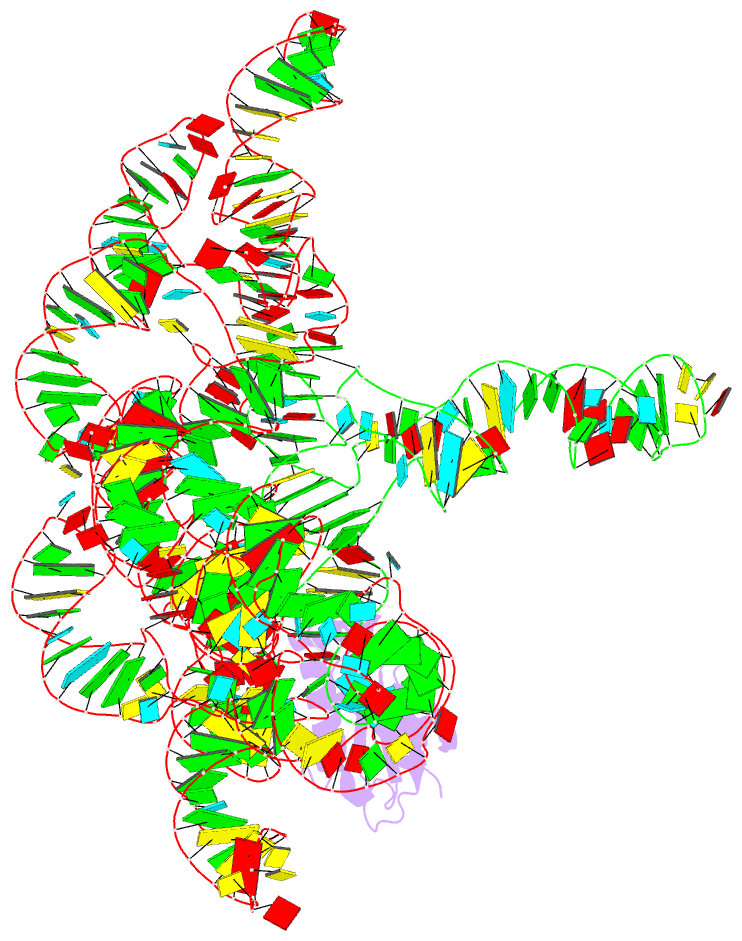
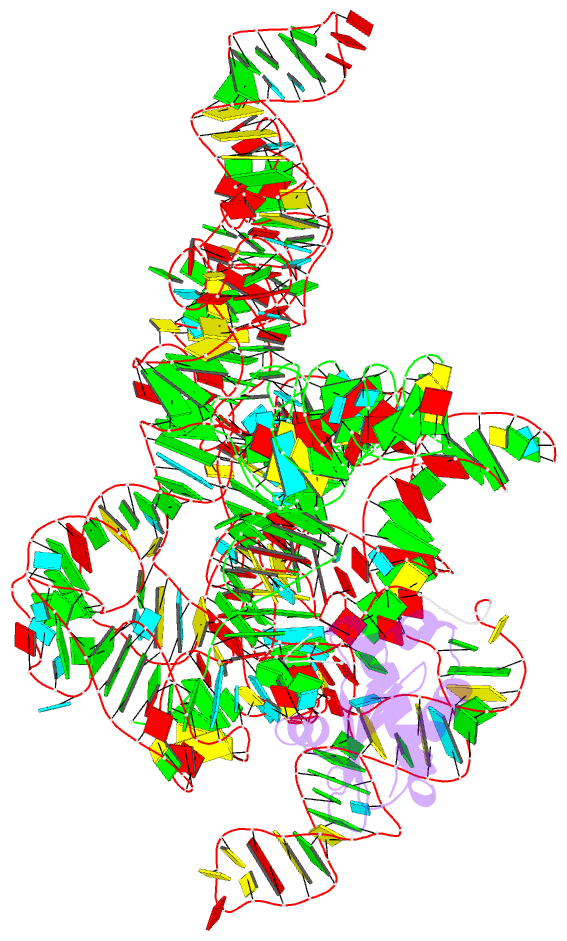
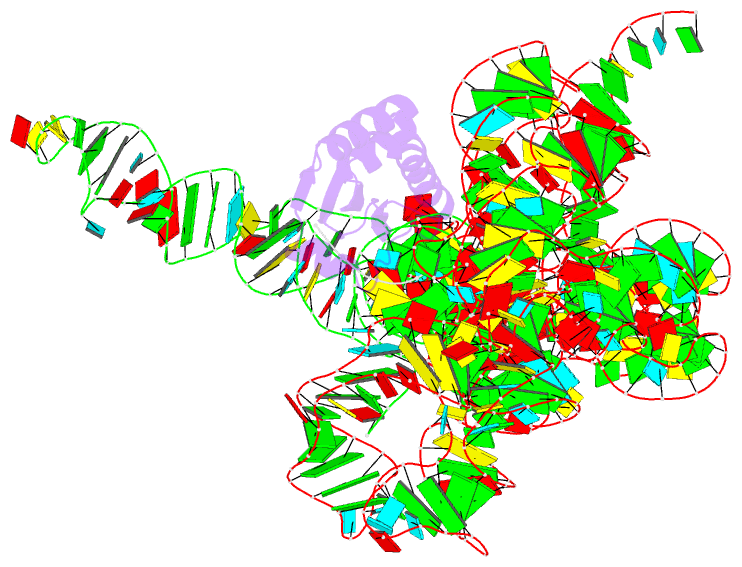
Summary information and primary citation
- PDB-id
- 3q1r; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (4.21 Å)
- Summary
- Crystal structure of a bacterial rnase p holoenzyme in complex with trna and in the presence of 5' leader
- Reference
- Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragon A (2010): "Structure of a Bacterial Ribonuclease P Holoenzyme in Complex with tRNA." Nature, 468, 784-789. doi: 10.1038/NATURE09516.
- Abstract
- Ribonuclease (RNase) P is the universal ribozyme responsible for 5'-end tRNA processing. We report the crystal structure of the Thermotoga maritima RNase P holoenzyme in complex with tRNA(Phe). The 154 kDa complex consists of a large catalytic RNA (P RNA), a small protein cofactor and a mature tRNA. The structure shows that RNA-RNA recognition occurs through shape complementarity, specific intermolecular contacts and base-pairing interactions. Soaks with a pre-tRNA 5' leader sequence with and without metal help to identify the 5' substrate path and potential catalytic metal ions. The protein binds on top of a universally conserved structural module in P RNA and interacts with the leader, but not with the mature tRNA. The active site is composed of phosphate backbone moieties, a universally conserved uridine nucleobase, and at least two catalytically important metal ions. The active site structure and conserved RNase P-tRNA contacts suggest a universal mechanism of catalysis by RNase P.