Summary information and primary citation
- PDB-id
- 3qsu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- chaperone-RNA
- Method
- X-ray (2.2 Å)
- Summary
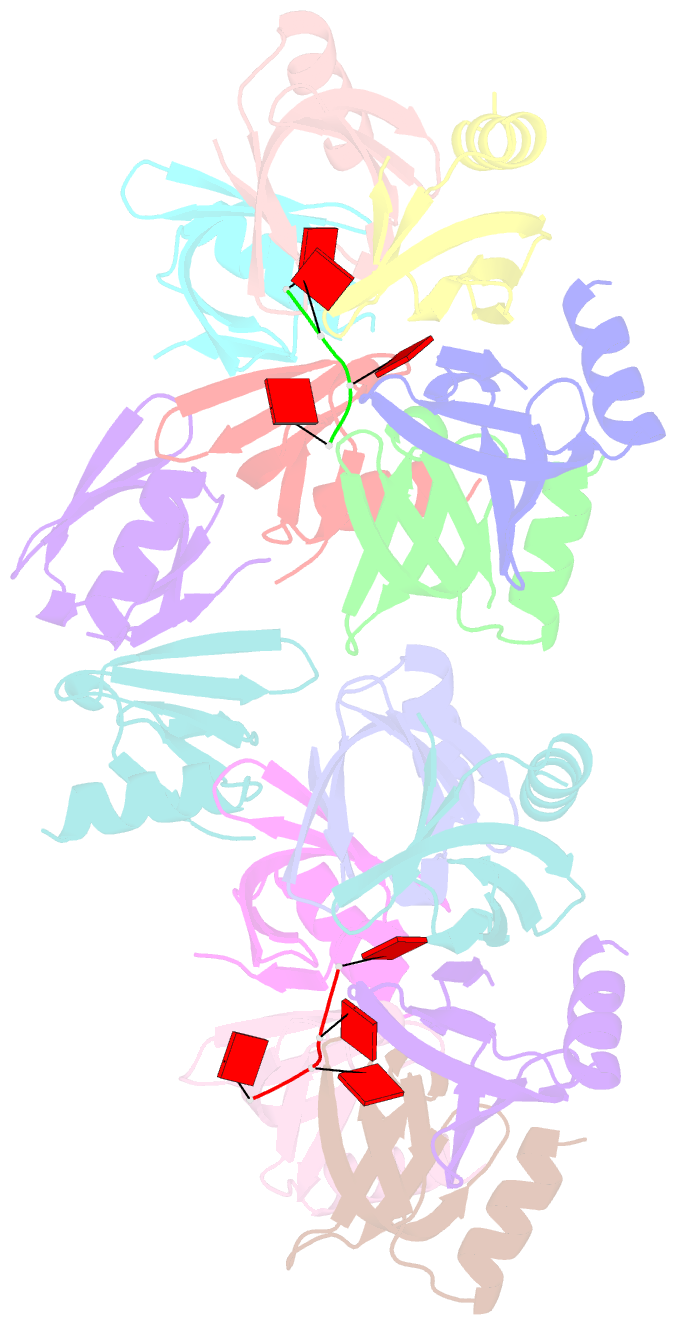
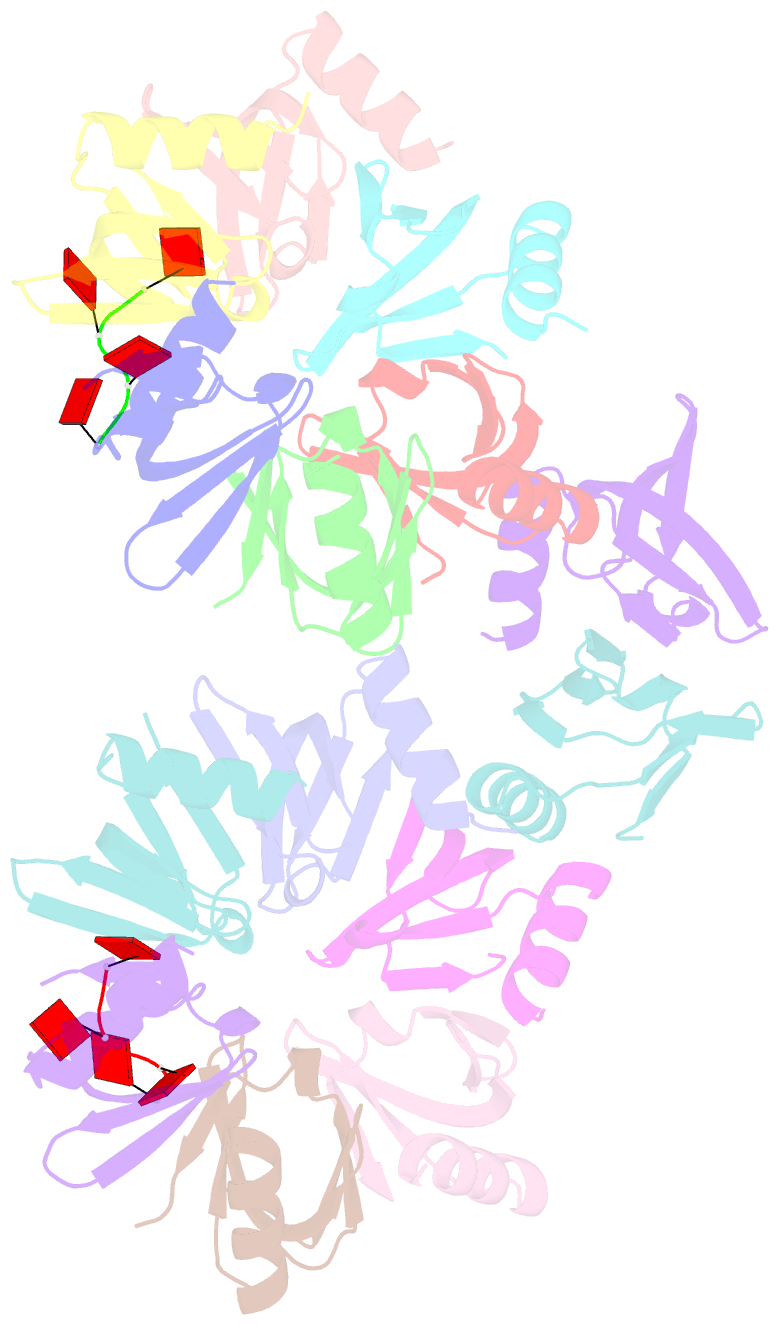
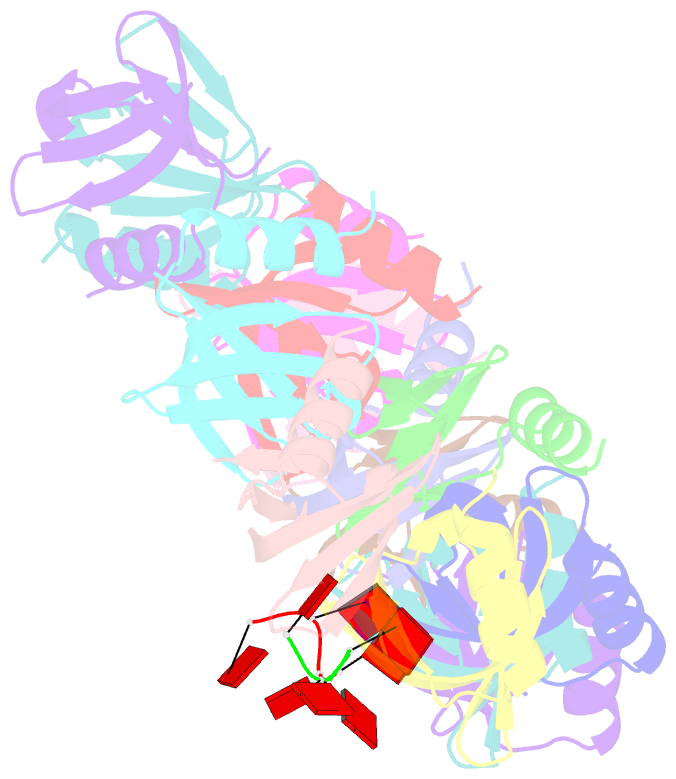
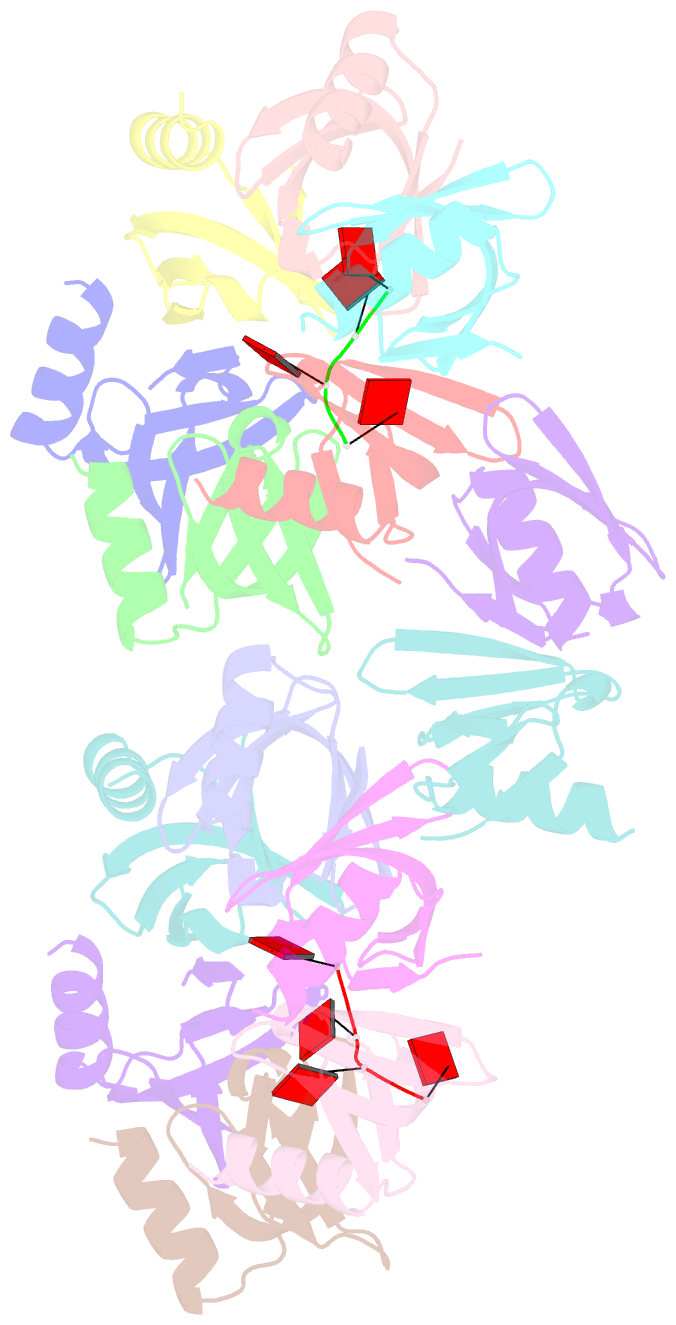
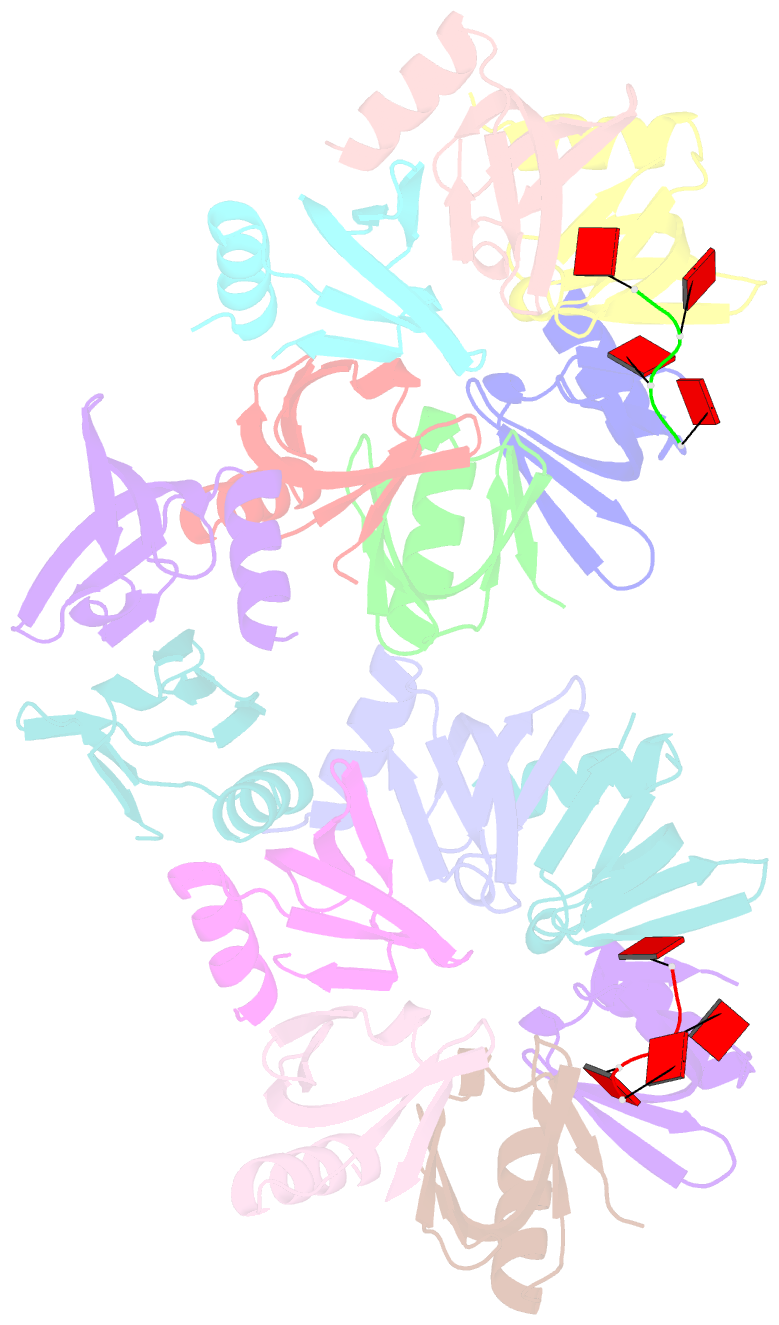
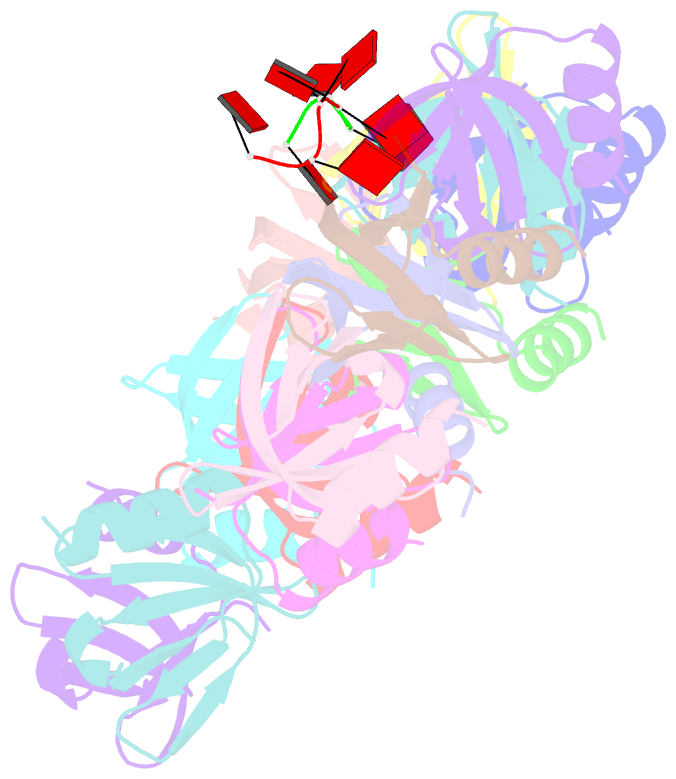
- Structure of staphylococcus aureus hfq in complex with a7 RNA
- Reference
- Horstmann N, Orans J, Valentin-Hansen P, Shelburne SA, Brennan RG (2012): "Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract." Nucleic Acids Res., 40, 11023-11035. doi: 10.1093/nar/gks809.
- Abstract
- Hfq is a post-transcriptional regulator that plays a key role in bacterial gene expression by binding AU-rich sequences and A-tracts to facilitate the annealing of sRNAs to target mRNAs and to affect RNA stability. To understand how Hfq from the Gram-positive bacterium Staphylococcus aureus (Sa) binds A-tract RNA, we determined the crystal structure of an Sa Hfq-adenine oligoribonucleotide complex. The structure reveals a bipartite RNA-binding motif on the distal face that is composed of a purine nucleotide-specificity site (R-site) and a non-discriminating linker site (L-site). The (R-L)-binding motif, which is also utilized by Bacillus subtilis Hfq to bind (AG)(3)A, differs from the (A-R-N) tripartite poly(A) RNA-binding motif of Escherichia coli Hfq whereby the Sa Hfq R-site strongly prefers adenosine, is more aromatic and permits deeper insertion of the adenine ring. R-site adenine-stacking residue Phe30, which is conserved among Gram-positive bacterial Hfqs, and an altered conformation about β3 and β4 eliminate the adenosine-specificity site (A-site) and create the L-site. Binding studies show that Sa Hfq binds (AU)(3)A ≈ (AG)(3)A ≥ (AC)(3)A > (AA)(3)A and L-site residue Lys33 plays a significant role. The (R-L) motif is likely utilized by Hfqs from most Gram-positive bacteria to bind alternating (A-N)(n) RNA.