Summary information and primary citation
- PDB-id
- 3r2d; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-RNA
- Method
- X-ray (2.199 Å)
- Summary
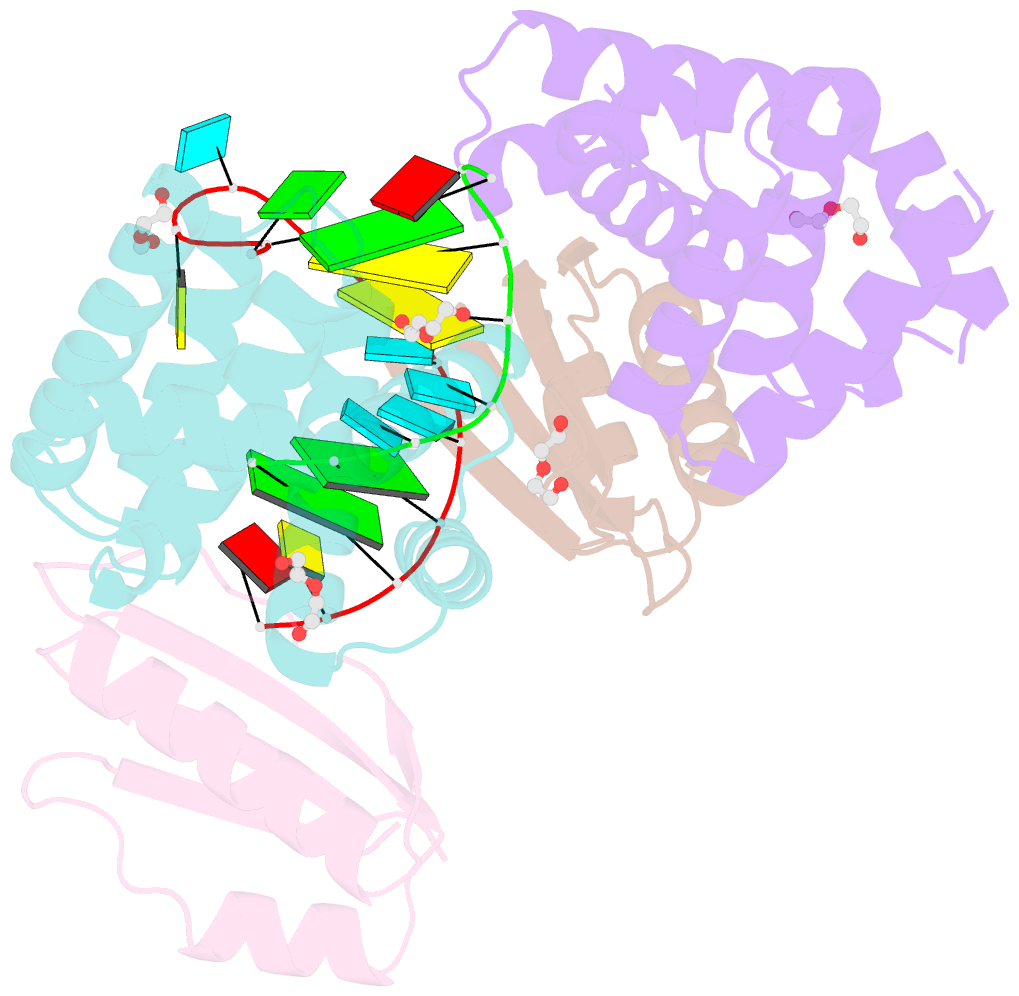
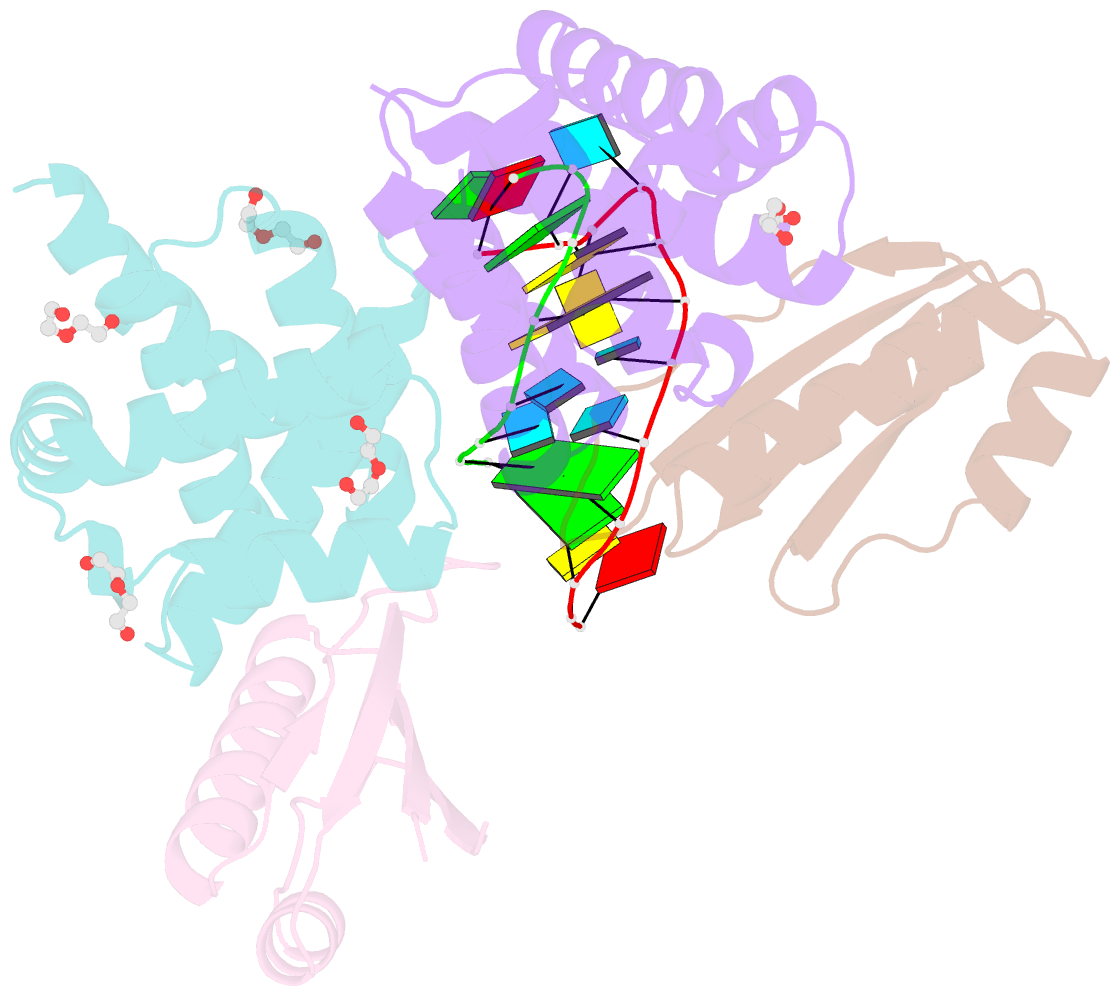
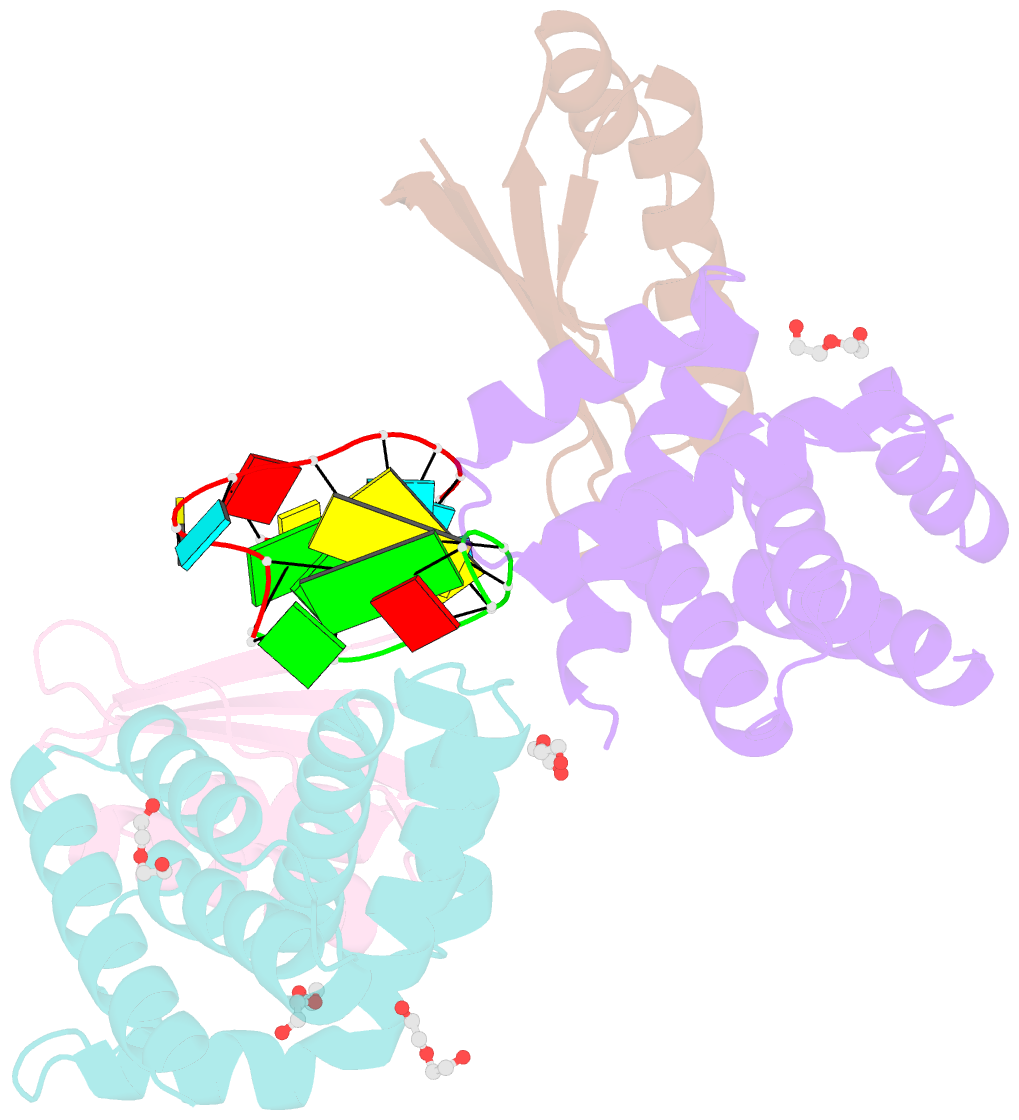
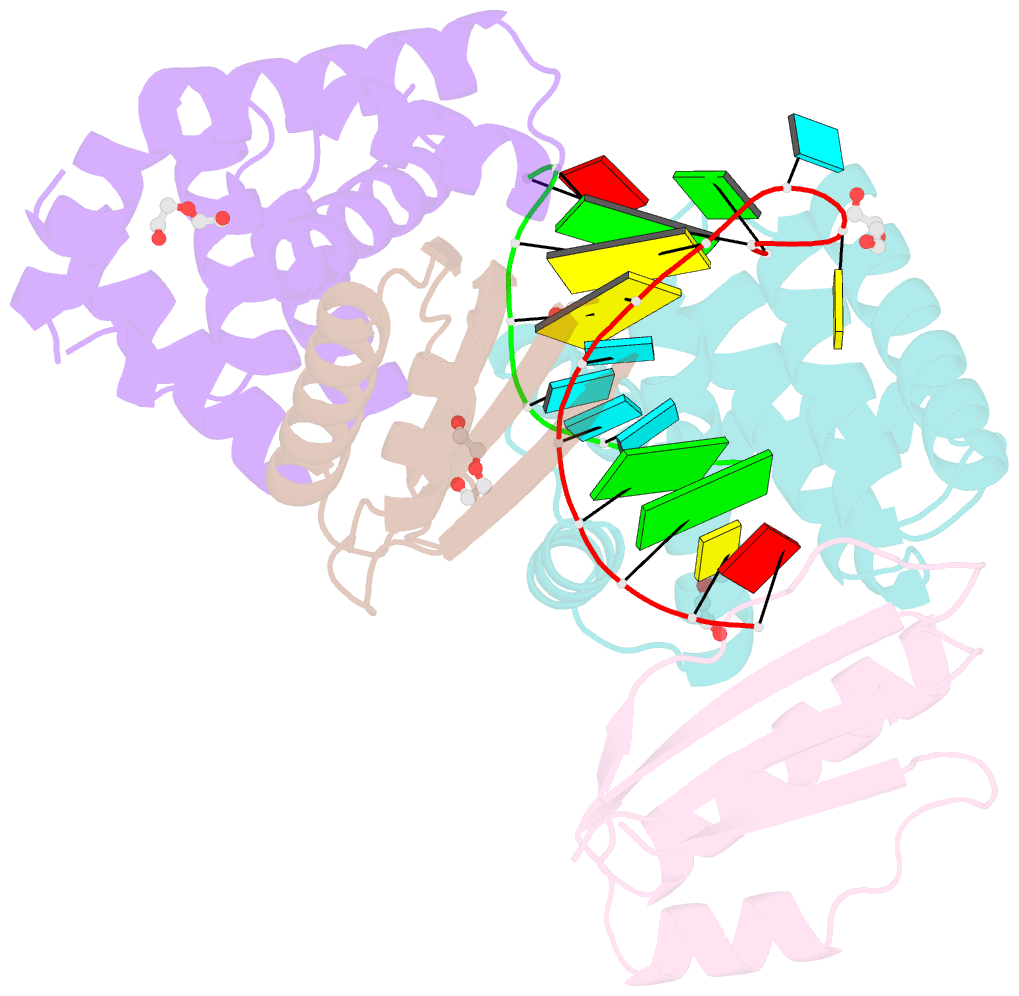
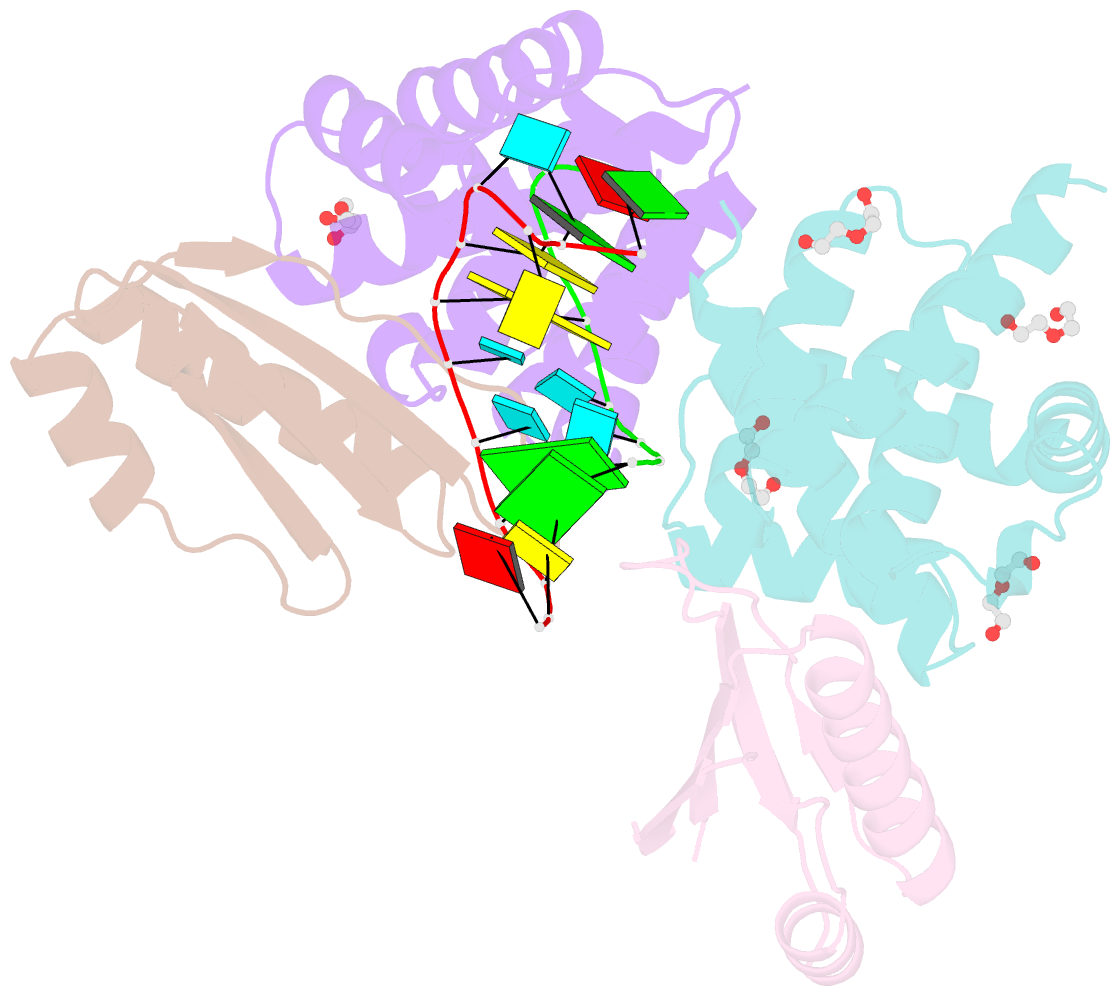
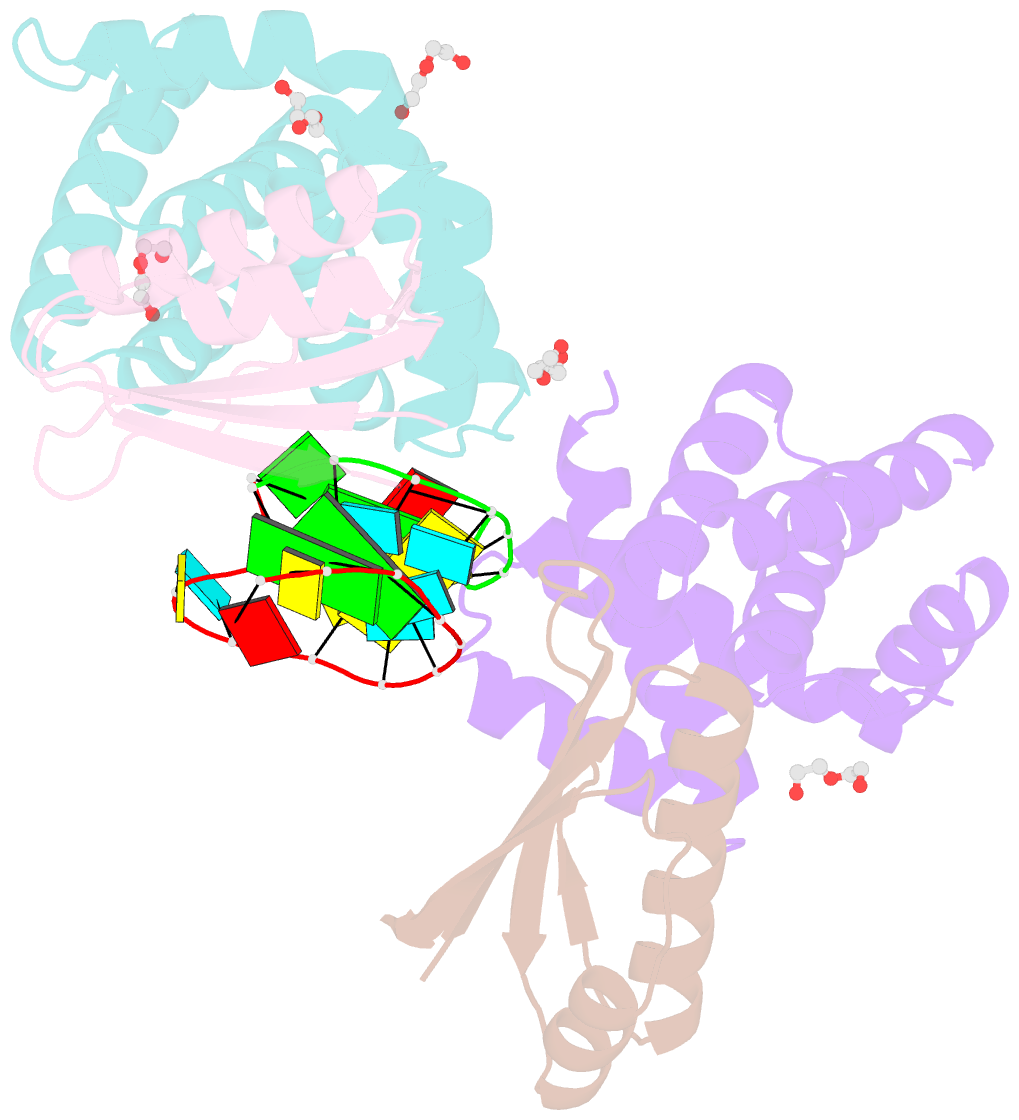
- Crystal structure of antitermination factors nusb and nuse in complex with dsrna
- Reference
- Stagno JR, Altieri AS, Bubunenko M, Tarasov SG, Li J, Court DL, Byrd RA, Ji X (2011): "Structural basis for RNA recognition by NusB and NusE in the initiation of transcription antitermination." Nucleic Acids Res., 39, 7803-7815. doi: 10.1093/nar/gkr418.
- Abstract
- Processive transcription antitermination requires the assembly of the complete antitermination complex, which is initiated by the formation of the ternary NusB-NusE-BoxA RNA complex. We have elucidated the crystal structure of this complex, demonstrating that the BoxA RNA is composed of 8 nt that are recognized by the NusB-NusE heterodimer. Functional biologic and biophysical data support the structural observations and establish the relative significance of key protein-protein and protein-RNA interactions. Further crystallographic investigation of a NusB-NusE-dsRNA complex reveals a heretofore unobserved dsRNA binding site contiguous with the BoxA binding site. We propose that the observed dsRNA represents BoxB RNA, as both single-stranded BoxA and double-stranded BoxB components are present in the classical lambda antitermination site. Combining these data with known interactions amongst antitermination factors suggests a specific model for the assembly of the complete antitermination complex.