Summary information and primary citation
- PDB-id
- 3r9x; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-transferase-RNA
- Method
- X-ray (2.8 Å)
- Summary
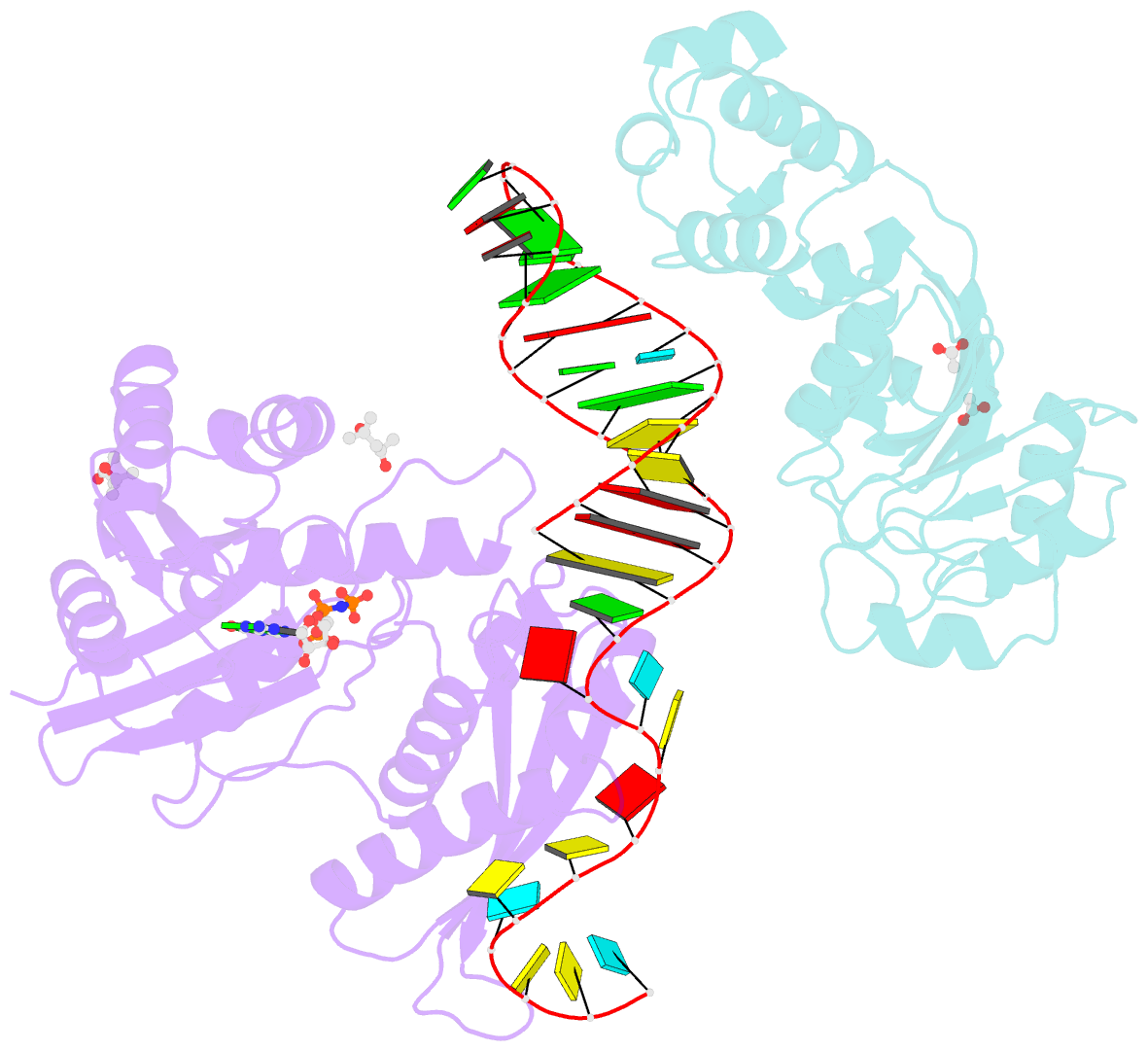
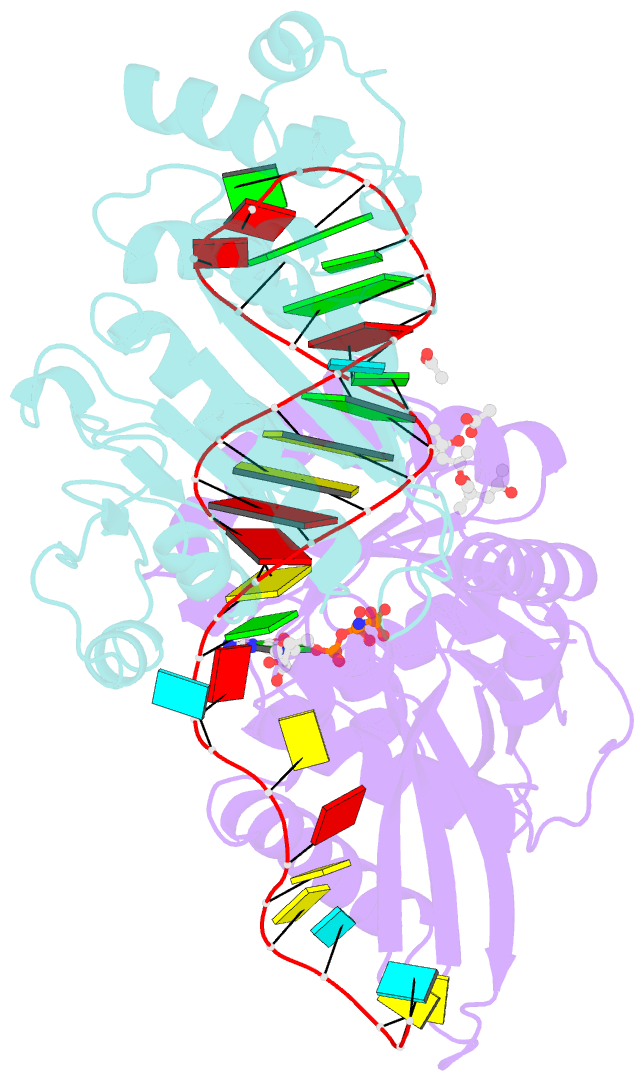
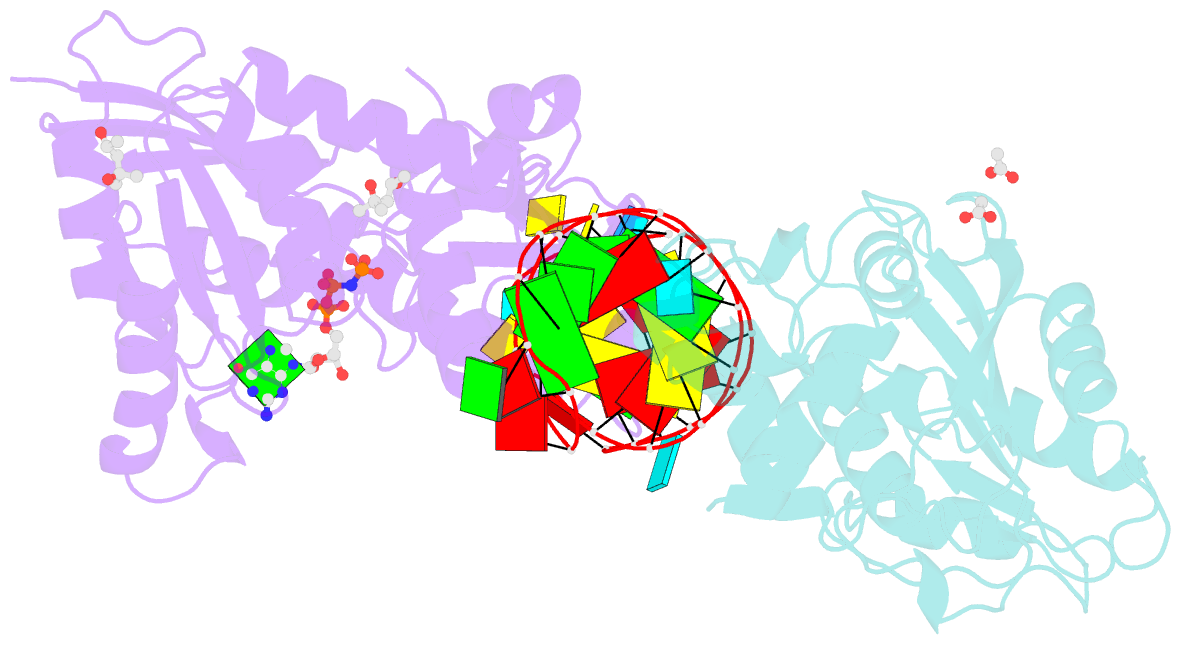
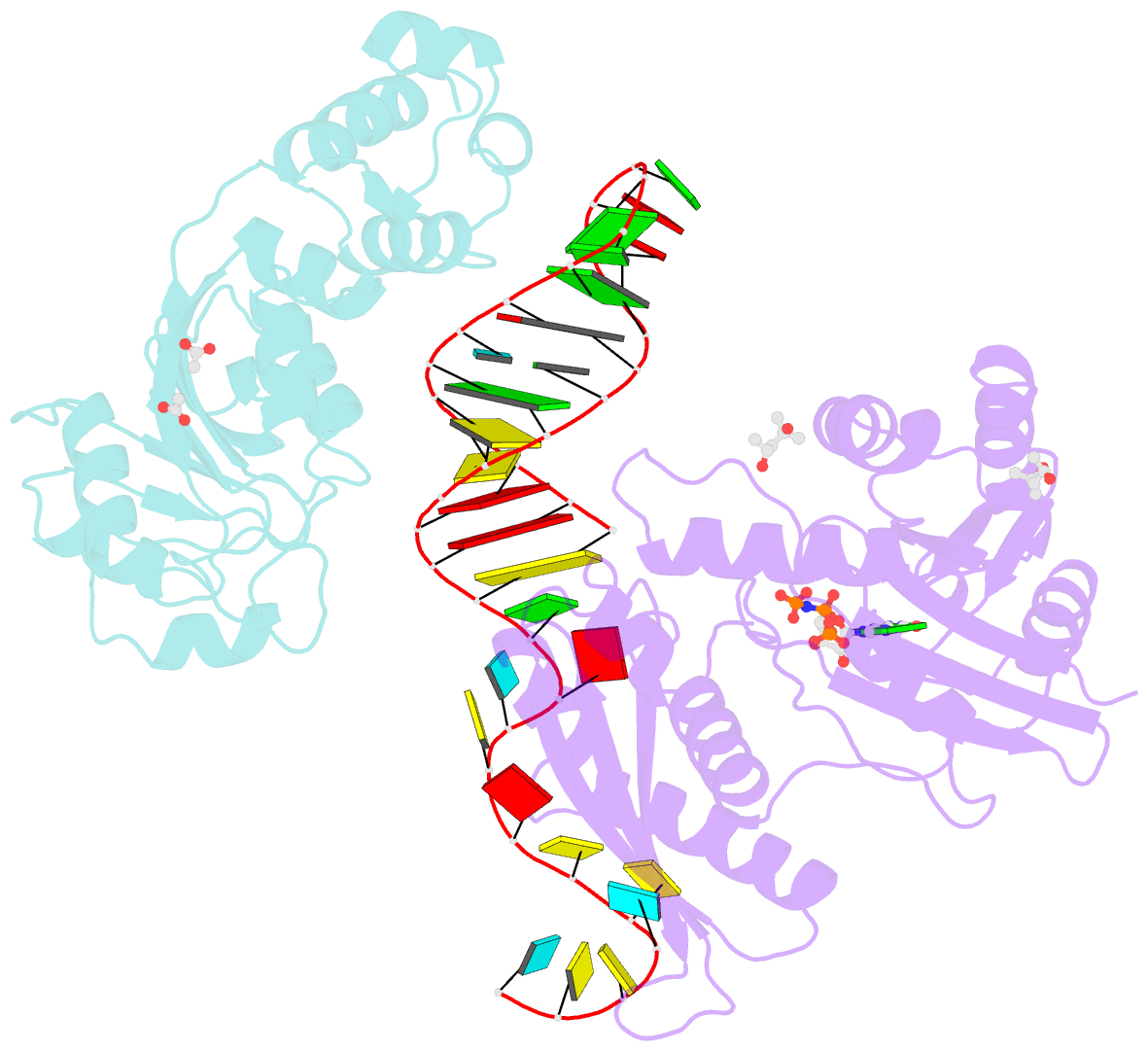
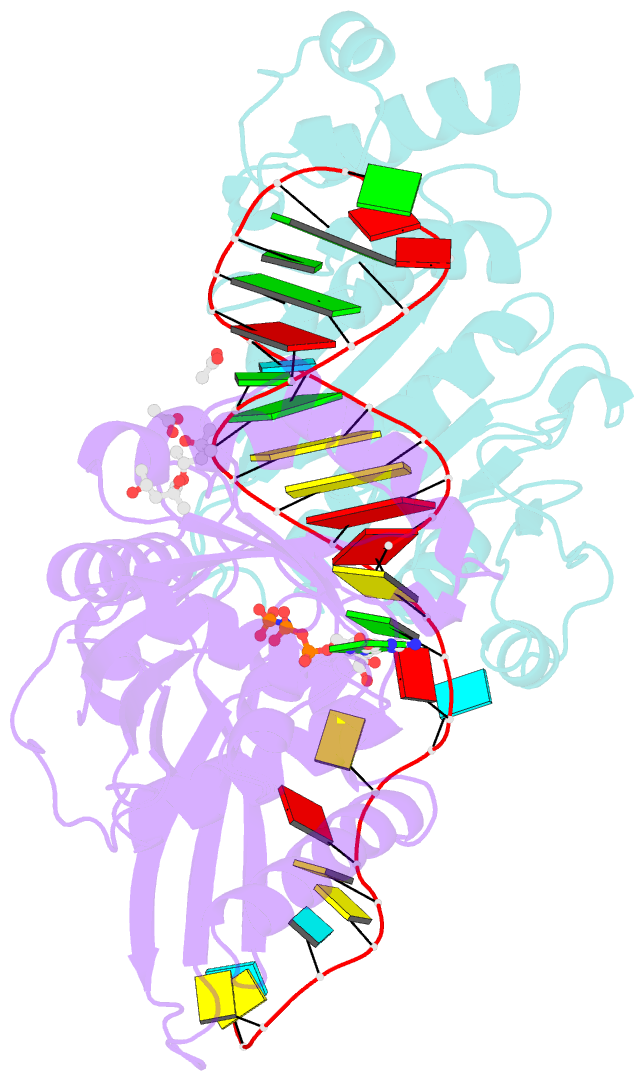
- Crystal structure of era in complex with mggdpnp, nucleotides 1506-1542 of 16s ribosomal RNA, and ksga
- Reference
- Tu C, Zhou X, Tarasov SG, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X (2011): "The Era GTPase recognizes the GAUCACCUCC sequence and binds helix 45 near the 3' end of 16S rRNA." Proc.Natl.Acad.Sci.USA, 108, 10156-10161. doi: 10.1073/pnas.1017679108.
- Abstract
- Era, composed of a GTPase domain and a K homology domain, is essential for bacterial cell viability. It is required for the maturation of 16S rRNA and assembly of the 30S ribosomal subunit. We showed previously that the protein recognizes nine nucleotides ( ) near the 3' end of 16S rRNA, and that this recognition stimulates GTP-hydrolyzing activity of Era. In all three kingdoms of life, the sequence and helix 45 (h45) (nucleotides 1506-1529) are highly conserved. It has been shown that the to double mutation severely affects the viability of bacteria. However, whether Era interacts with G1530 and/or h45 and whether such interactions (if any) contribute to the stimulation of Era's GTPase activity were not known. Here, we report two RNA structures that contain nucleotides 1506-1542 (RNA301), one in complex with Era and GDPNP (GNP), a nonhydrolysable GTP-analogue, and the other in complex with Era, GNP, and the KsgA methyltransferase. The structures show that Era recognizes 10 nucleotides, including G1530, and that Era also binds h45. Moreover, GTPase assay experiments show that G1530 does not stimulate Era's GTPase activity. Rather, A1531 and A1534 are most important for stimulation and h45 further contributes to the stimulation. Although G1530 does not contribute to the intrinsic GTPase activity of Era, its interaction with Era is important for binding and is essential for the protein to function, leading to the discovery of a new cold-sensitive phenotype of Era.