Summary information and primary citation
- PDB-id
- 3rc8; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.9 Å)
- Summary
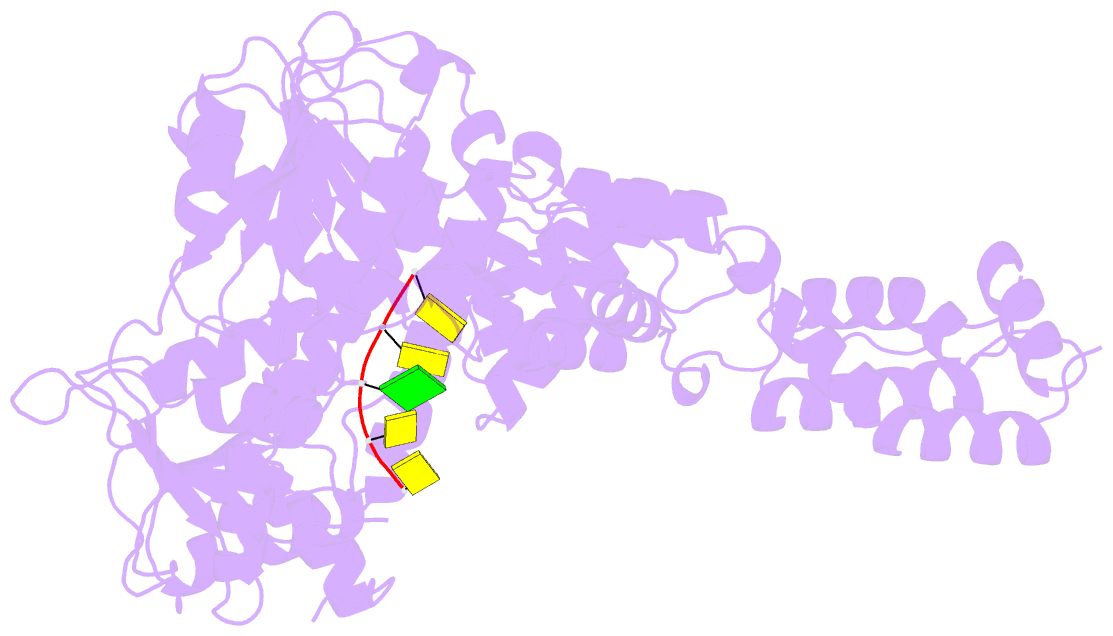
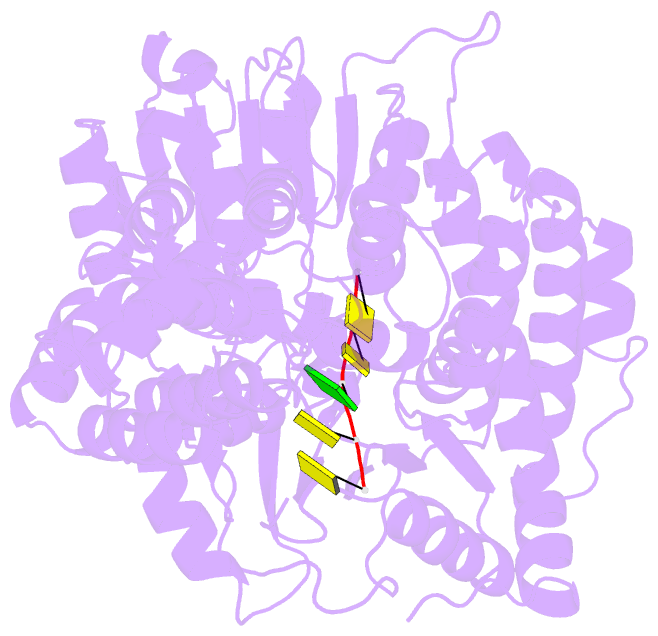
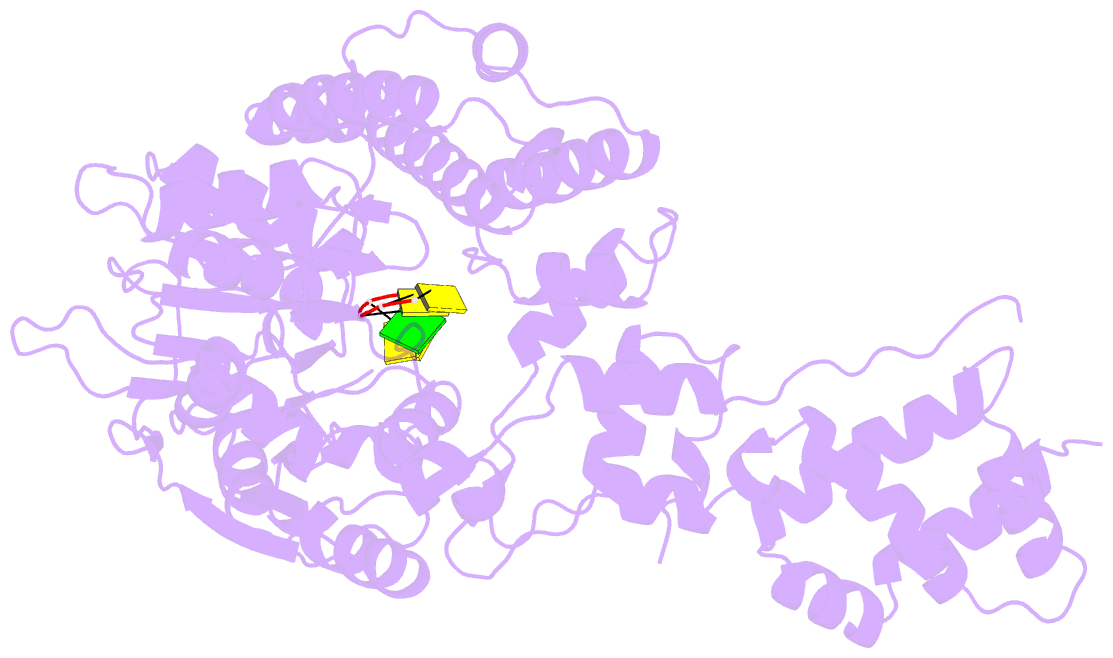
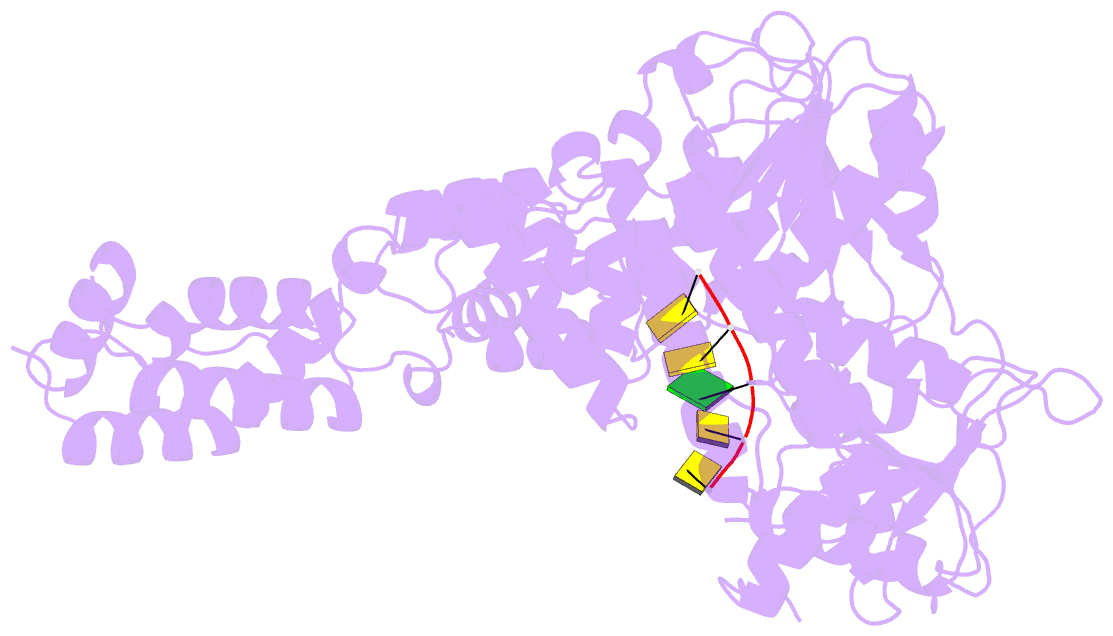
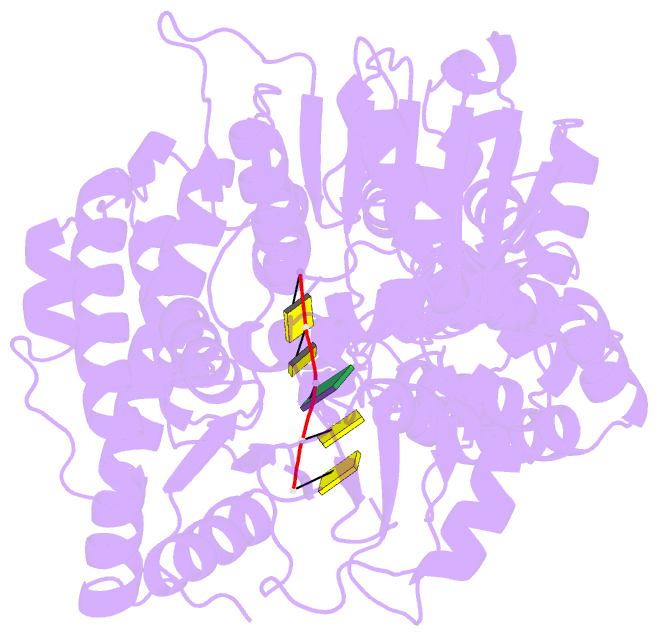
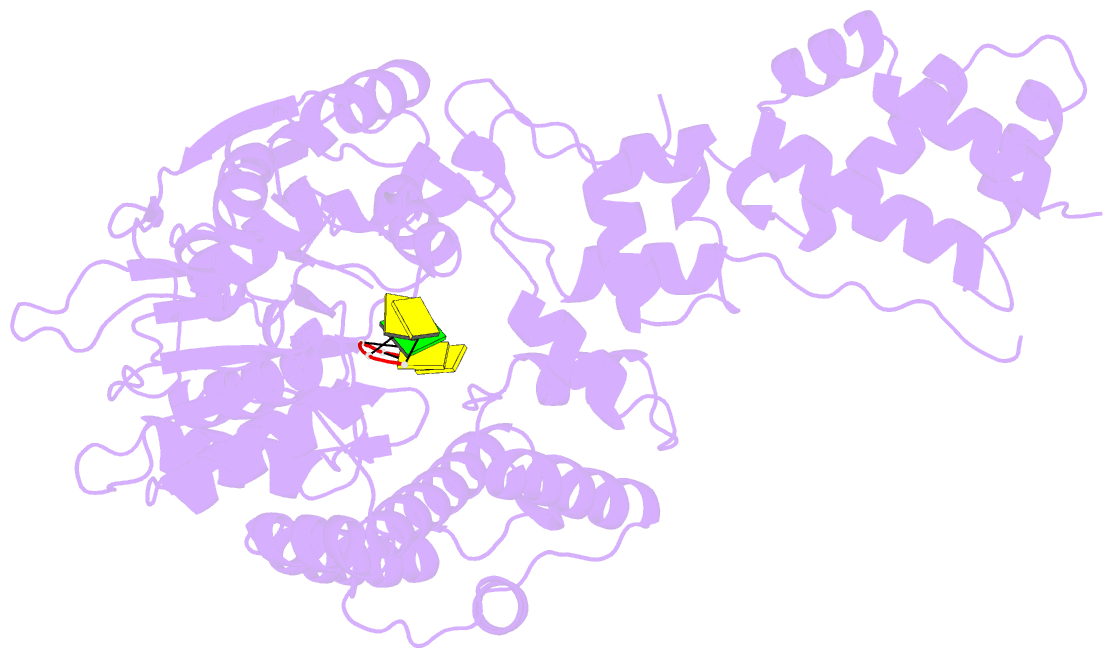
- Human mitochondrial helicase suv3 in complex with short RNA fragment
- Reference
- Jedrzejczak R, Wang J, Dauter M, Szczesny RJ, Stepien PP, Dauter Z (2011): "Human Suv3 protein reveals unique features among SF2 helicases." Acta Crystallogr.,Sect.D, 67, 988-996. doi: 10.1107/S0907444911040248.
- Abstract
- Suv3 is a helicase that is involved in efficient turnover and surveillance of RNA in eukaryotes. In vitro studies show that human Suv3 (hSuv3) in complex with human polynucleotide phosphorylase has RNA degradosome activity. The enzyme is mainly localized in mitochondria, but small fractions are found in cell nuclei. Here, two X-ray crystallographic structures of human Suv3 in complex with AMPPNP, a nonhydrolysable analog of ATP, and with a short five-nucleotide strand of RNA are presented at resolutions of 2.08 and 2.9 Å, respectively. The structure of the enzyme is very similar in the two complexes and consists of four domains. Two RecA-like domains form the tandem typical of all helicases from the SF2 superfamily which together with the C-terminal all-helical domain makes a ring structure through which the nucleotide strand threads. The mostly helical N-terminal domain is positioned externally with respect to the core of the enzyme. Most of the typical helicase motifs are present in hSuv3, but the protein shows certain unique characteristics, suggesting that Suv3 enzymes may constitute a separate subfamily of helicases.