Summary information and primary citation
- PDB-id
- 3rel; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- structural protein-DNA
- Method
- X-ray (2.7 Å)
- Summary
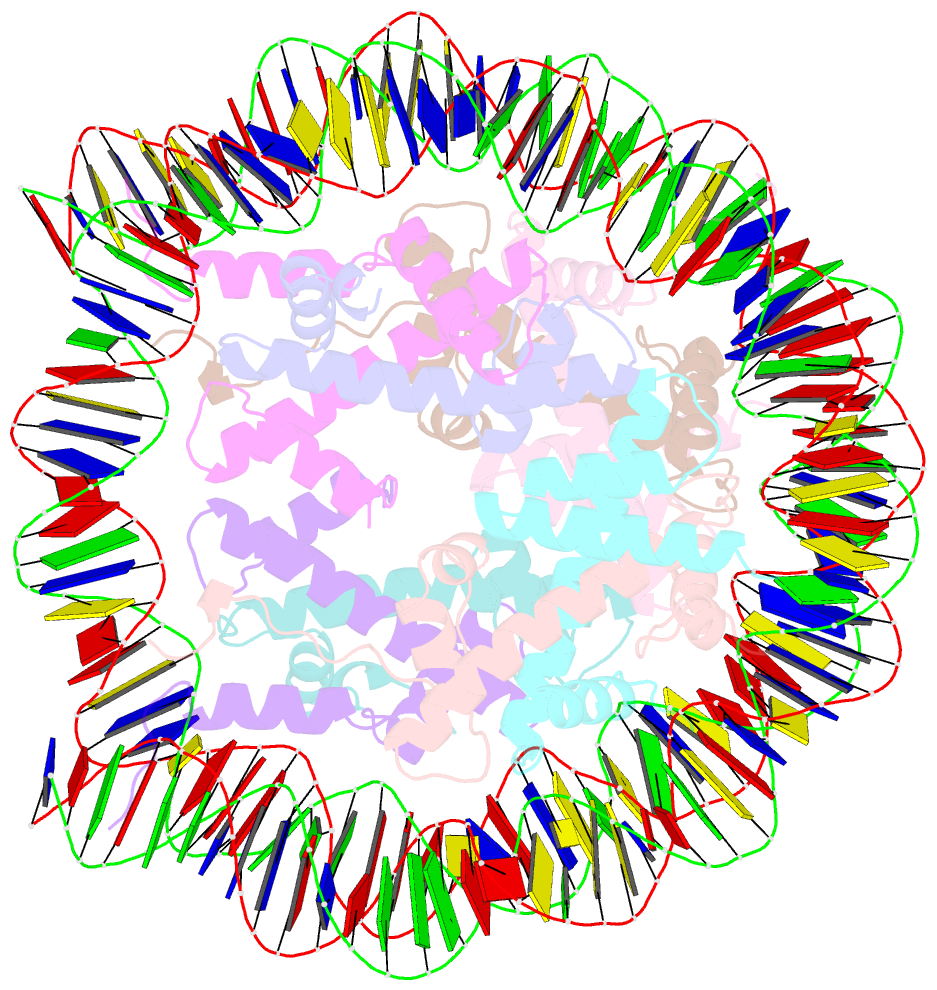
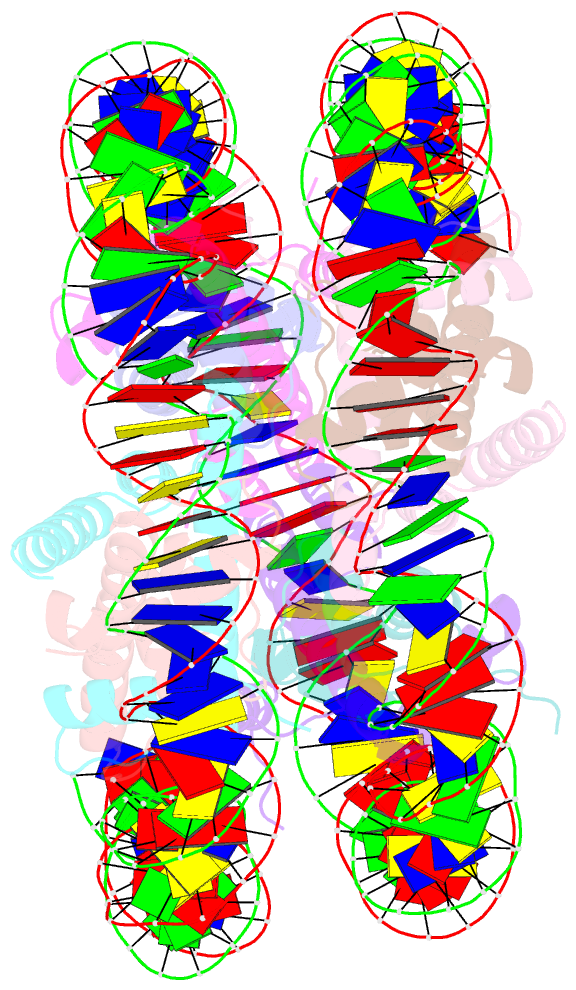
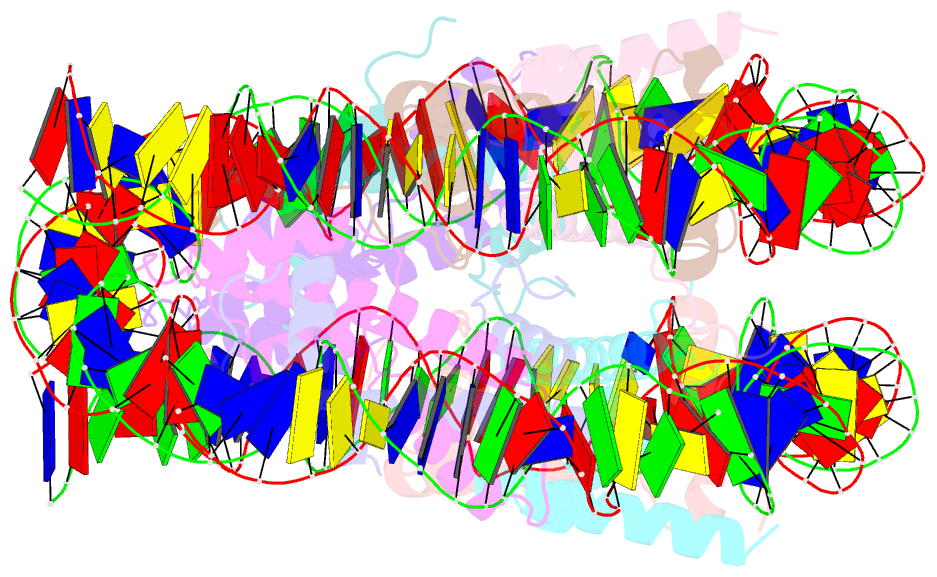
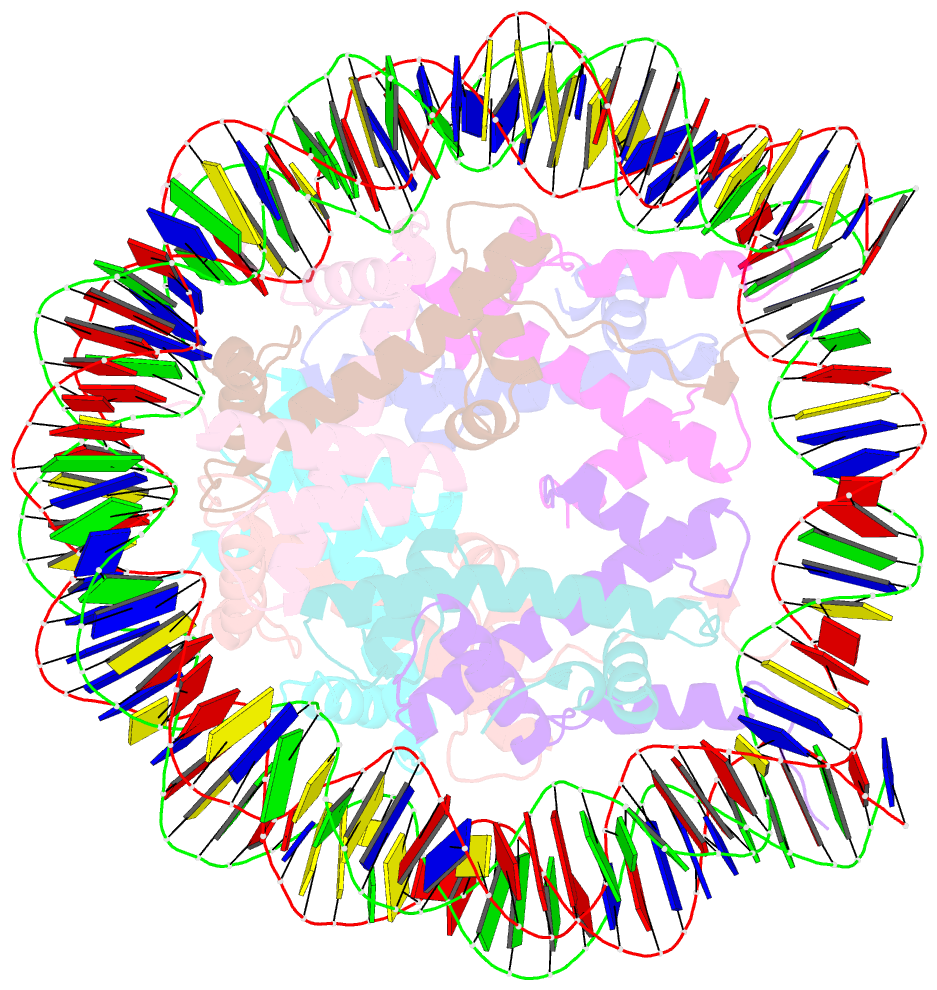
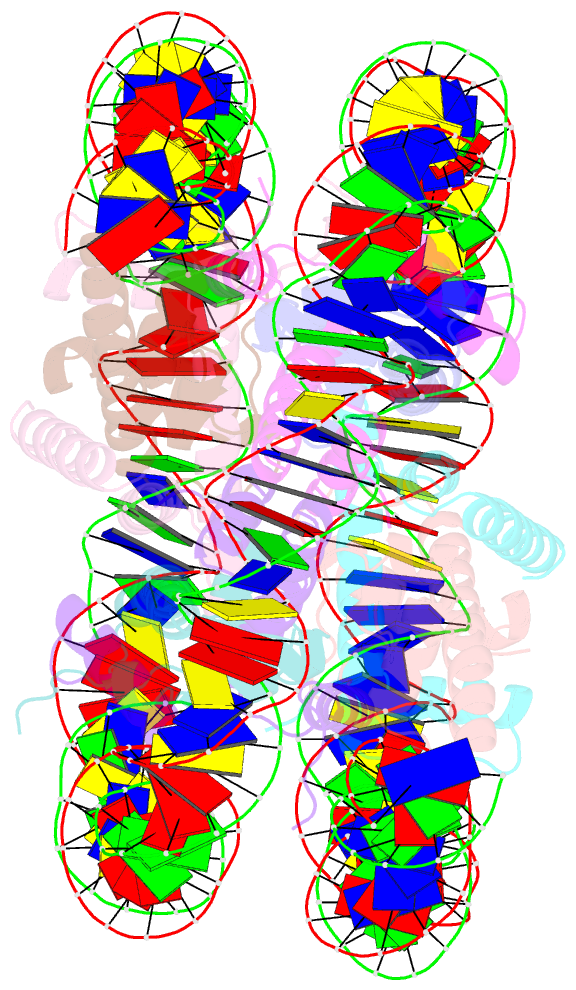
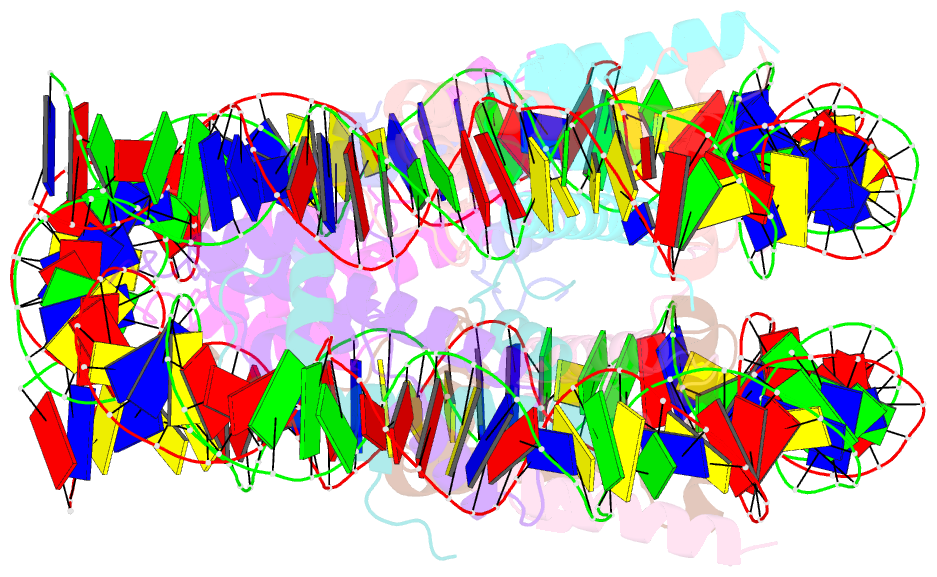
- 2.7 angstrom crystal structure of the nucleosome core particle assembled with a 146 bp alpha-satellite DNA (ncp146b) derivatized with triamminechloroplatinum(ii) chloride
- Reference
- Wu B, Davey GE, Nazarov AA, Dyson PJ, Davey CA (2011): "Specific DNA structural attributes modulate platinum anticancer drug site selection and cross-link generation." Nucleic Acids Res., 39, 8200-8212. doi: 10.1093/nar/gkr491.
- Abstract
- Heavy metal compounds have toxic and medicinal potential through capacity to form strong specific bonds with macromolecules, and the interaction of platinum drugs at the major groove nitrogen atom of guanine bases primarily underlies their therapeutic activity. By crystallographic analysis of transition metal-and in particular platinum compound-DNA site selectivity in the nucleosome core, we establish that steric accessibility, which is controlled by specific structural parameters of the double helix, modulates initial guanine-metal bond formation. Moreover, DNA conformational features can be linked to both similarities and distinctions in platinum drug adduct formation between the naked and nucleosomal DNA states. Notably, structures that facilitate initial platinum-guanine bond formation can oppose cross-link generation, rationalizing the occurrence of long-lived therapeutically ineffective monofunctional adducts. These findings illuminate DNA structure-dependent reactivity and provide a novel framework for understanding metal-double helix interactions, which should facilitate the development of improved chromatin-targeting medicinal agents.