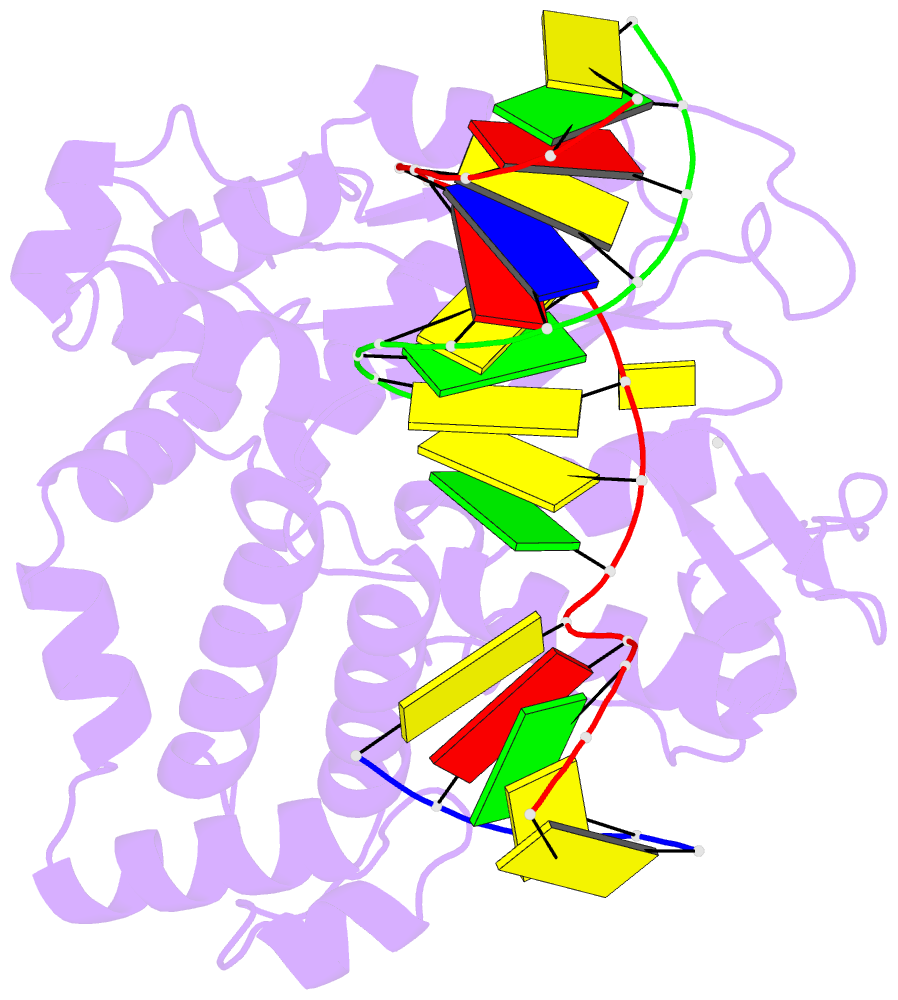
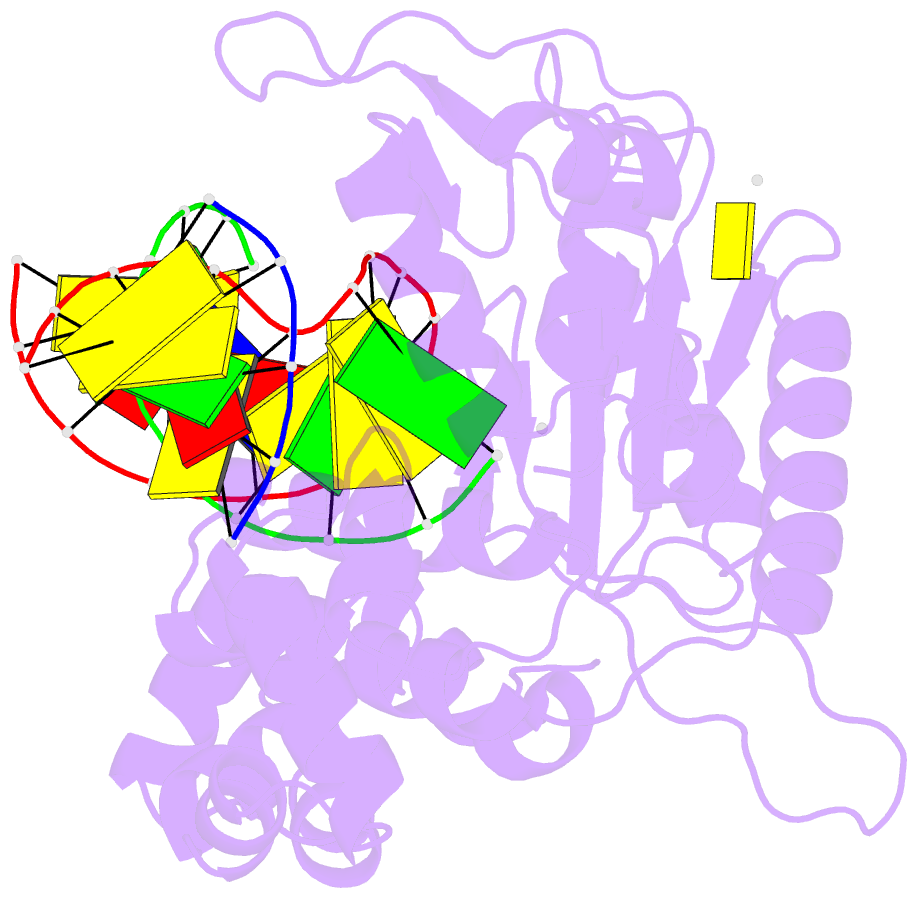
Summary information and primary citation
- PDB-id
- 3rh5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.096 Å)
- Summary
- DNA polymerase beta mutant (y271) with a dideoxy-terminated primer with an incoming deoxynucleotide (dctp)
- Reference
- Cavanaugh NA, Beard WA, Batra VK, Perera L, Pedersen LG, Wilson SH (2011): "Molecular insights into DNA polymerase deterrents for ribonucleotide insertion." J.Biol.Chem., 286, 31650-31660. doi: 10.1074/jbc.M111.253401.
- Abstract
- DNA polymerases can misinsert ribonucleotides that lead to genomic instability. DNA polymerase β discourages ribonucleotide insertion with the backbone carbonyl of Tyr-271; alanine substitution of Tyr-271, but not Phe-272, resulted in a >10-fold loss in discrimination. The Y271A mutant also inserted ribonucleotides more efficiently than wild type on a variety of ribonucleoside (rNMP)-containing DNA substrates. Substituting Mn(2+) for Mg(2+) decreased sugar discrimination for both wild-type and mutant enzymes primarily by increasing the affinity for rCTP. This facilitated crystallization of ternary substrate complexes of both the wild-type and Y271A mutant enzymes. Crystallographic structures of Y271A- and wild type-substrate complexes indicated that rCTP is well accommodated in the active site but that O2' of rCTP and the carbonyl oxygen of Tyr-271 or Ala-271 are unusually close (∼2.5 and 2.6 Å, respectively). Structure-based modeling indicates that the local energetic cost of positioning these closely spaced oxygens is ∼2.2 kcal/mol for the wild-type enzyme. Because the side chain of Tyr-271 also hydrogen bonds with the primer terminus, loss of this interaction affects its catalytic positioning. Our results support a model where DNA polymerase β utilizes two strategies, steric and geometric, with a single protein residue to deter ribonucleotide insertion.