Summary information and primary citation
- PDB-id
- 3sv3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.1 Å)
- Summary
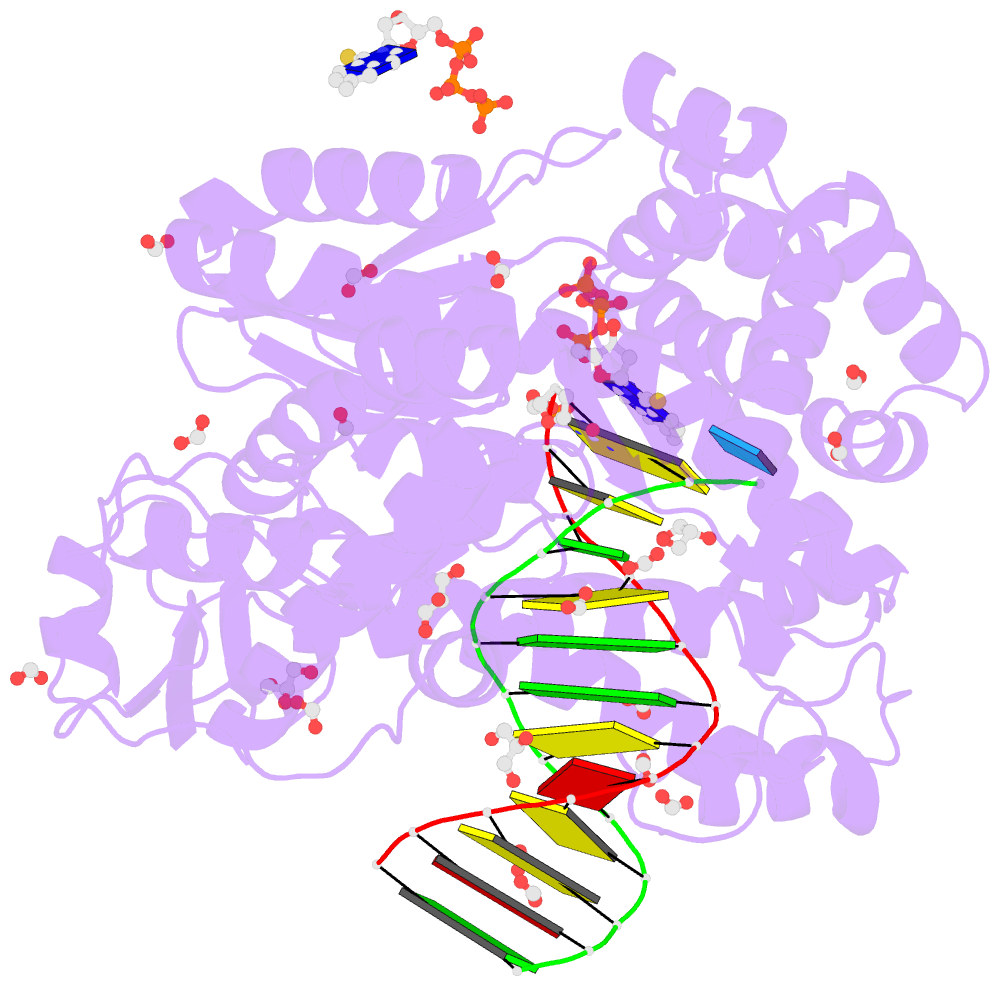
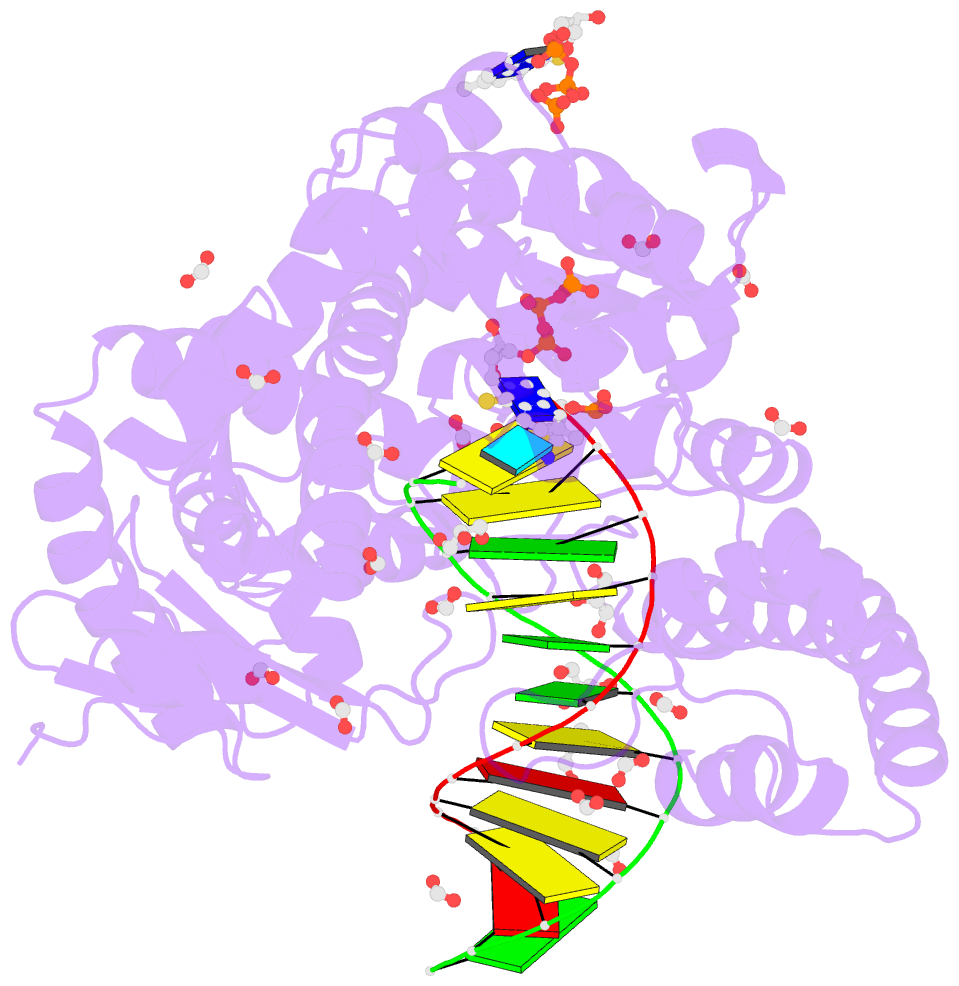
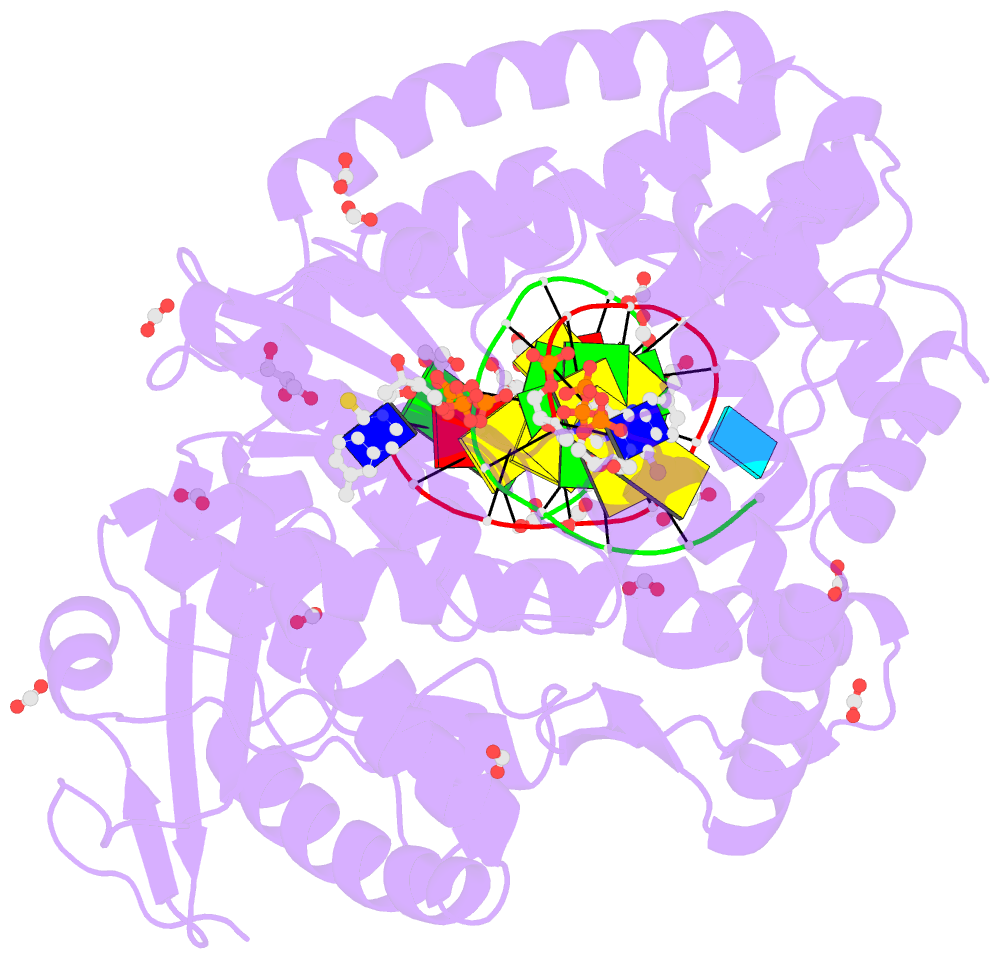
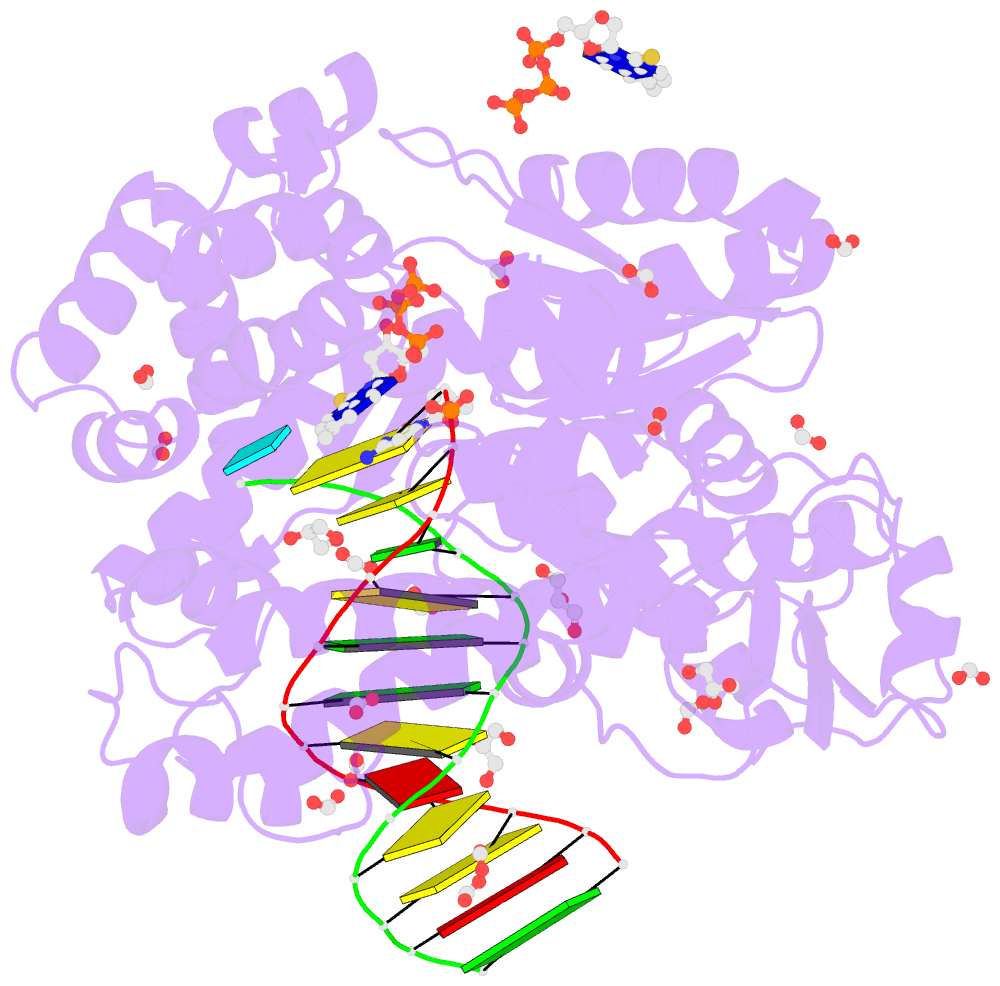
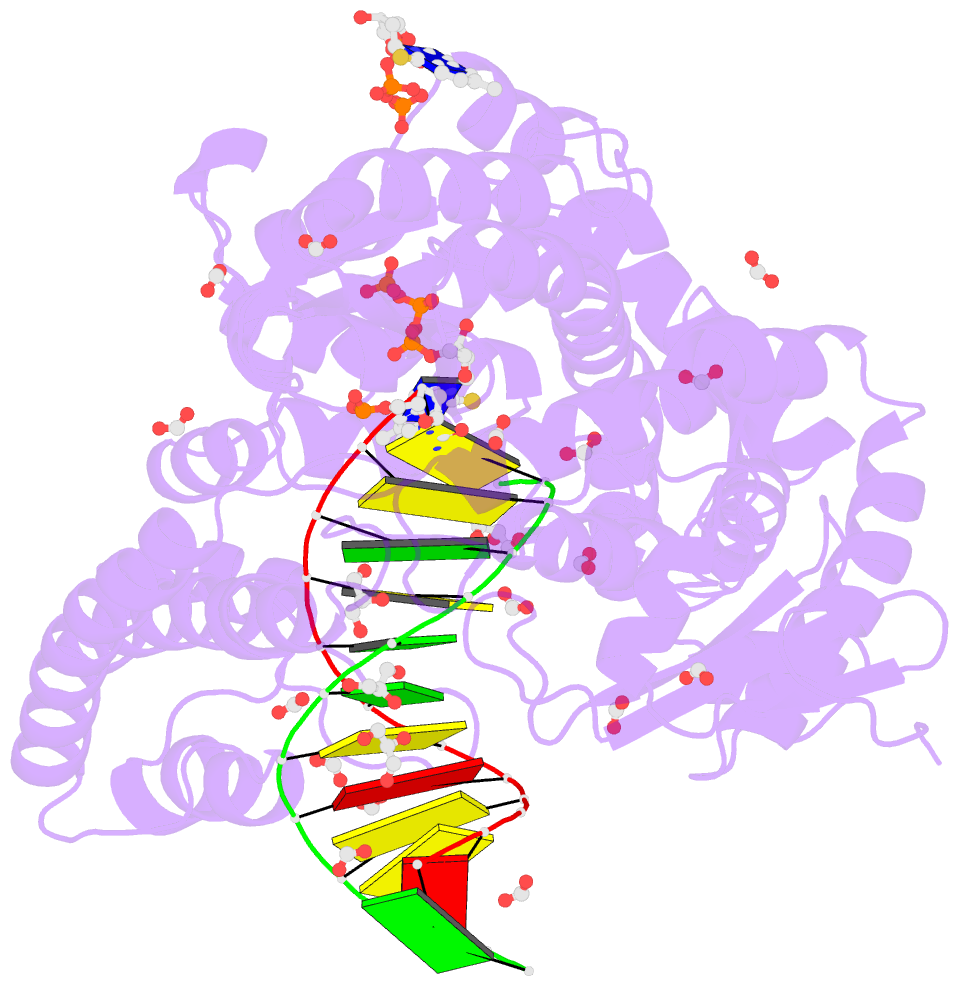
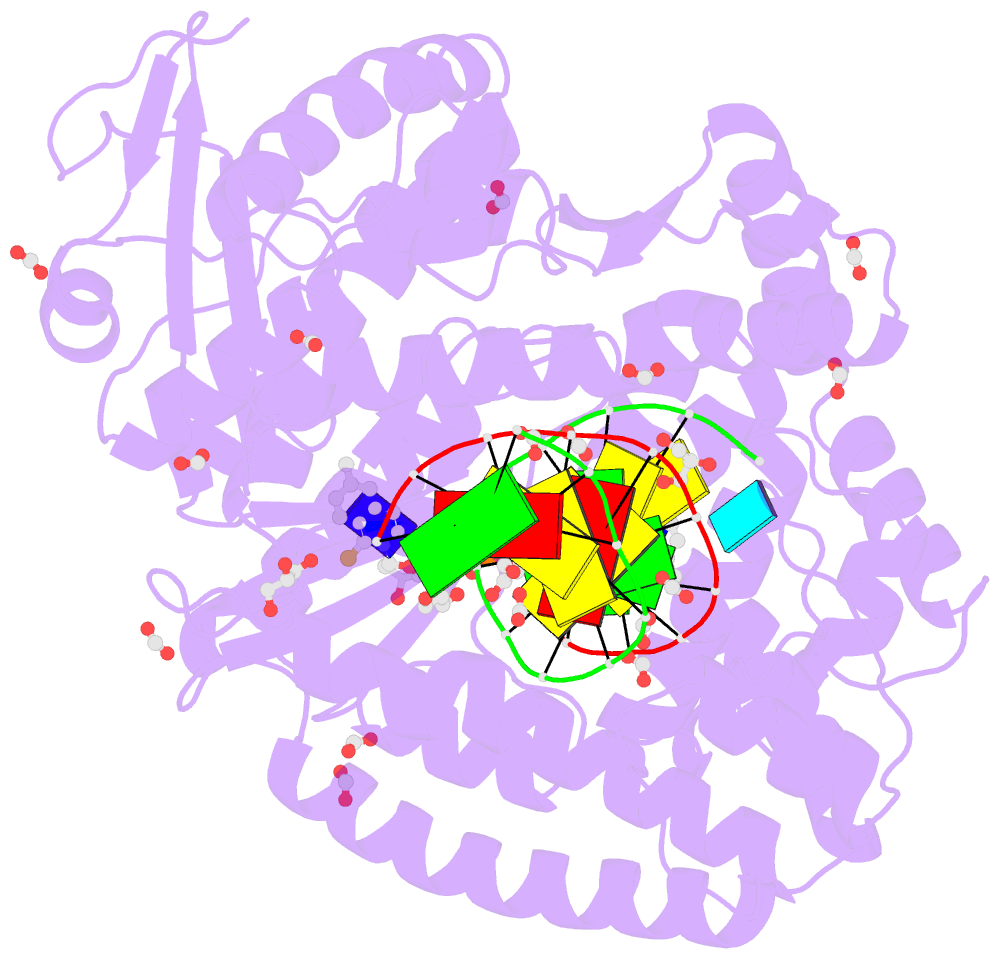
- Crystal structure of the large fragment of DNA polymerase i from thermus aquaticus in a closed ternary complex with the artificial base pair dnam-d5sicstp
- Reference
- Betz K, Malyshev DA, Lavergne T, Welte W, Diederichs K, Dwyer TJ, Ordoukhanian P, Romesberg FE, Marx A (2012): "KlenTaq polymerase replicates unnatural base pairs by inducing a Watson-Crick geometry." Nat.Chem.Biol., 8, 612-614. doi: 10.1038/nchembio.966.
- Abstract
- Many candidate unnatural DNA base pairs have been developed, but some of the best-replicated pairs adopt intercalated structures in free DNA that are difficult to reconcile with known mechanisms of polymerase recognition. Here we present crystal structures of KlenTaq DNA polymerase at different stages of replication for one such pair, dNaM-d5SICS, and show that efficient replication results from the polymerase itself, inducing the required natural-like structure.