Summary information and primary citation
- PDB-id
- 3szq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.353 Å)
- Summary
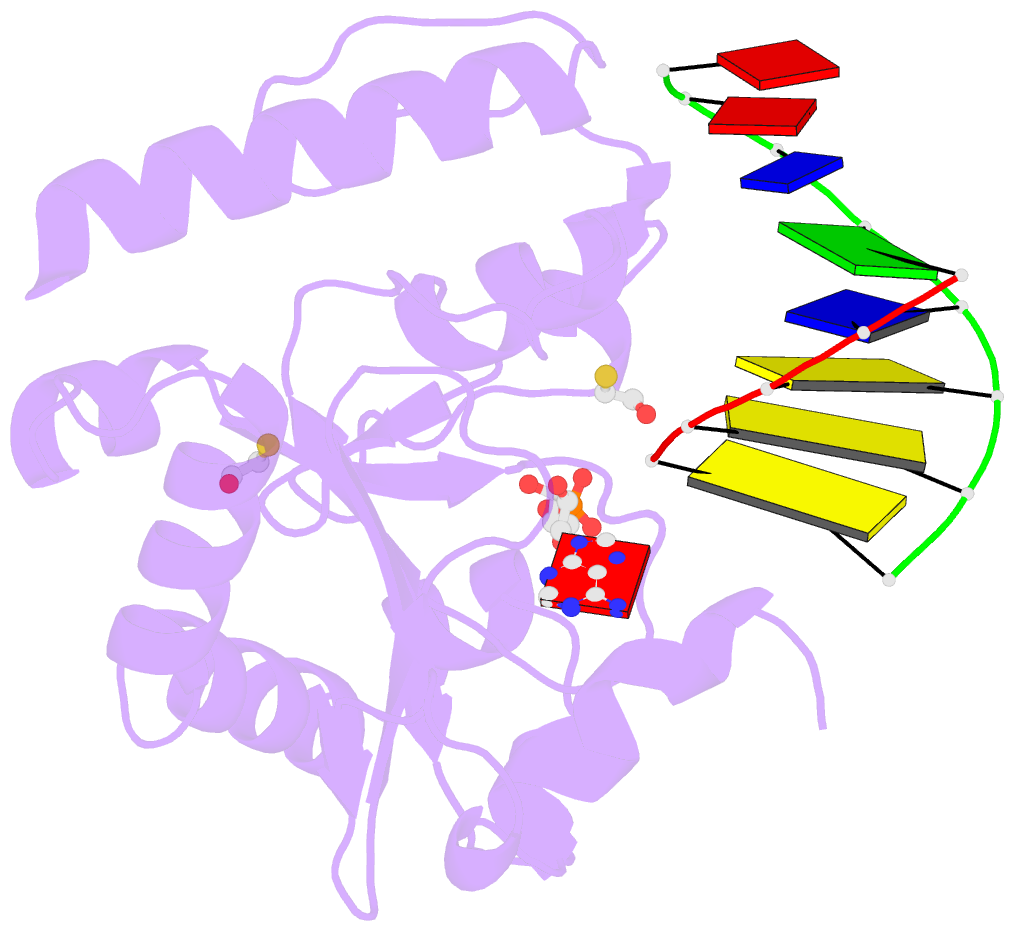
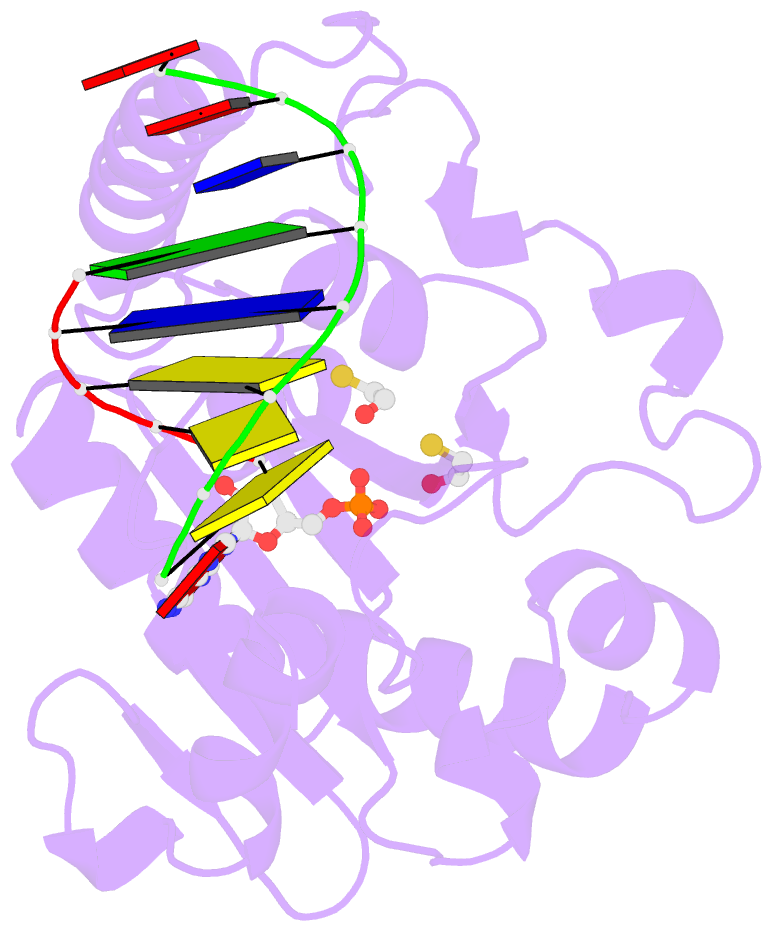
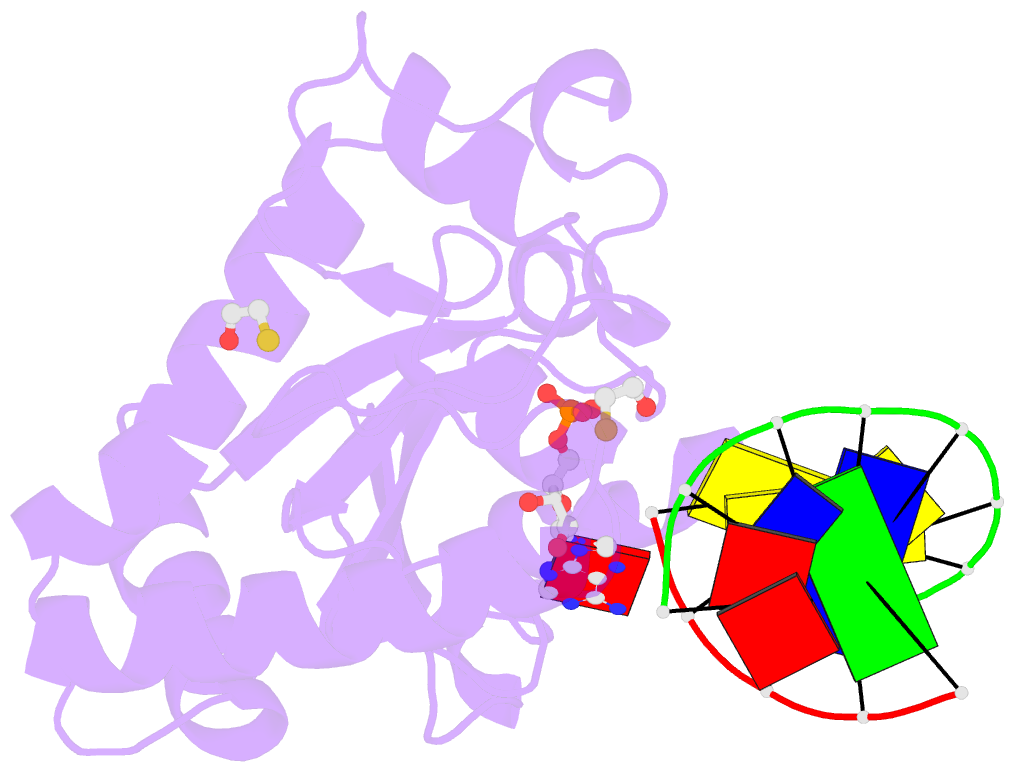
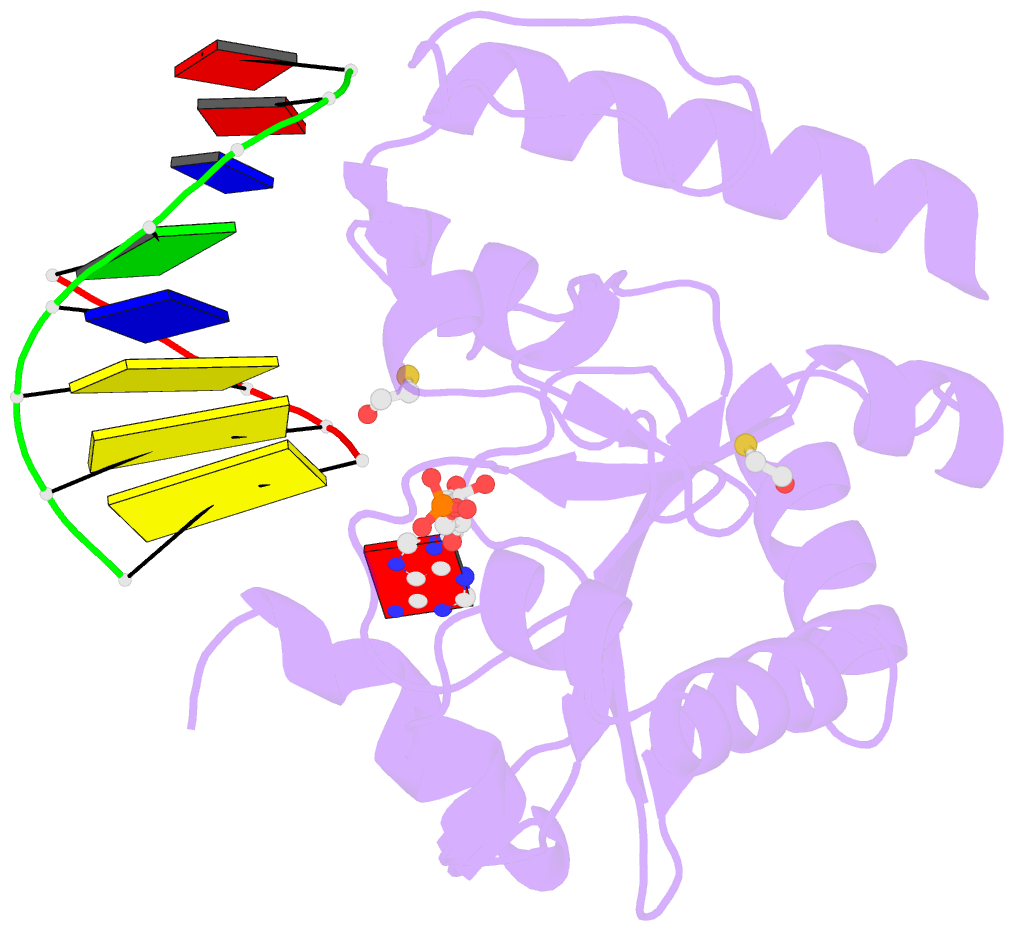
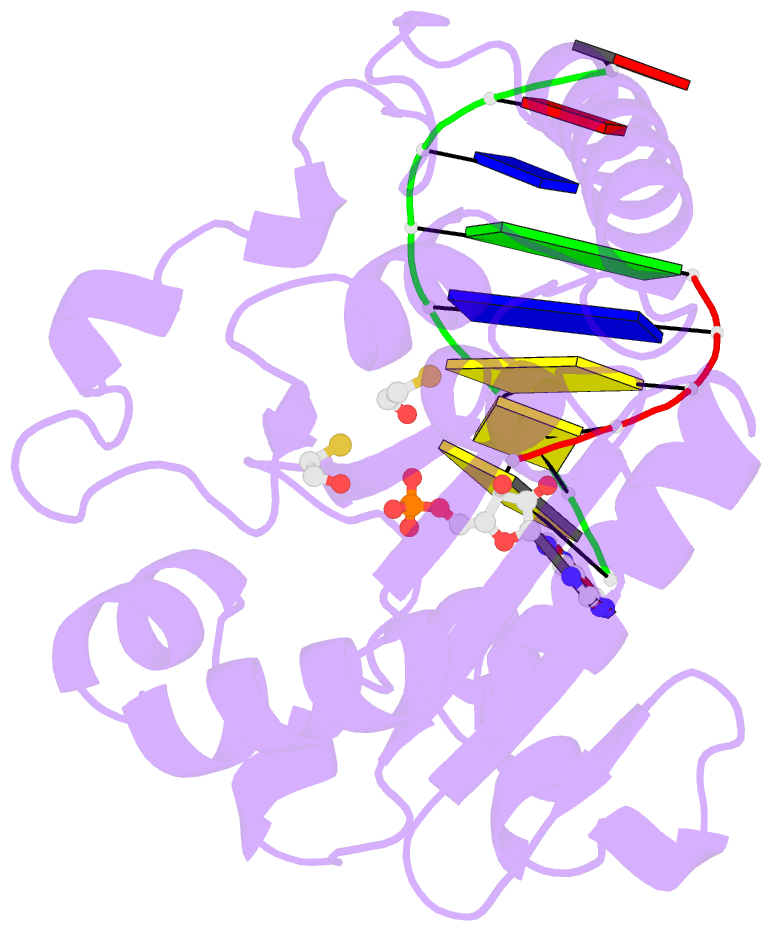
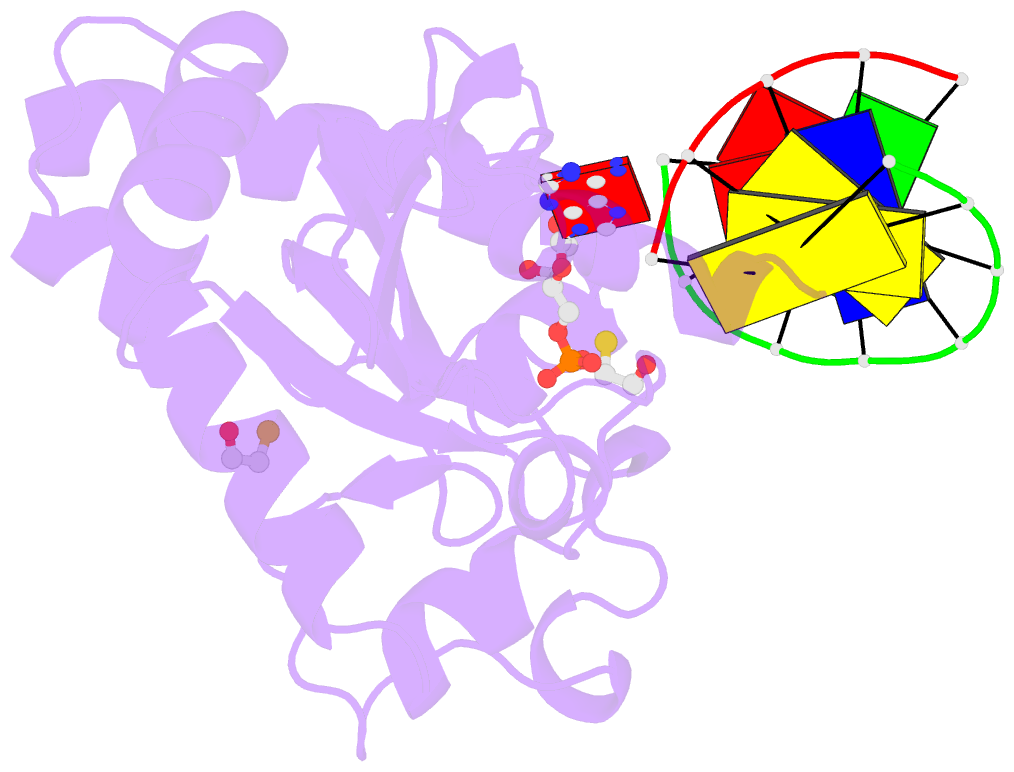
- Structure of an s. pombe aptx-DNA-amp-zn complex
- Reference
- Tumbale P, Appel CD, Kraehenbuehl R, Robertson PD, Williams JS, Krahn J, Ahel I, Williams RS (2011): "Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease." Nat.Struct.Mol.Biol., 18, 1189-1195. doi: 10.1038/nsmb.2146.
- Abstract
- DNA ligases finalize DNA replication and repair through DNA nick-sealing reactions that can abort to generate cytotoxic 5'-adenylation DNA damage. Aprataxin (Aptx) catalyzes direct reversal of 5'-adenylate adducts to protect genome integrity. Here the structure of a Schizosaccharomyces pombe Aptx-DNA-AMP-Zn(2+) complex reveals active site and DNA interaction clefts formed by fusing a histidine triad (HIT) nucleotide hydrolase with a DNA minor groove-binding C(2)HE zinc finger (Znf). An Aptx helical 'wedge' interrogates the base stack for sensing DNA ends or DNA nicks. The HIT-Znf, the wedge and an '[F/Y]PK' pivot motif cooperate to distort terminal DNA base-pairing and direct 5'-adenylate into the active site pocket. Structural and mutational data support a wedge-pivot-cut HIT-Znf catalytic mechanism for 5'-adenylate adduct recognition and removal and suggest that mutations affecting protein folding, the active site pocket and the pivot motif underlie Aptx dysfunction in the neurodegenerative disorder ataxia with oculomotor apraxia 1 (AOA1).