Summary information and primary citation
- PDB-id
- 3tq1; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.556 Å)
- Summary
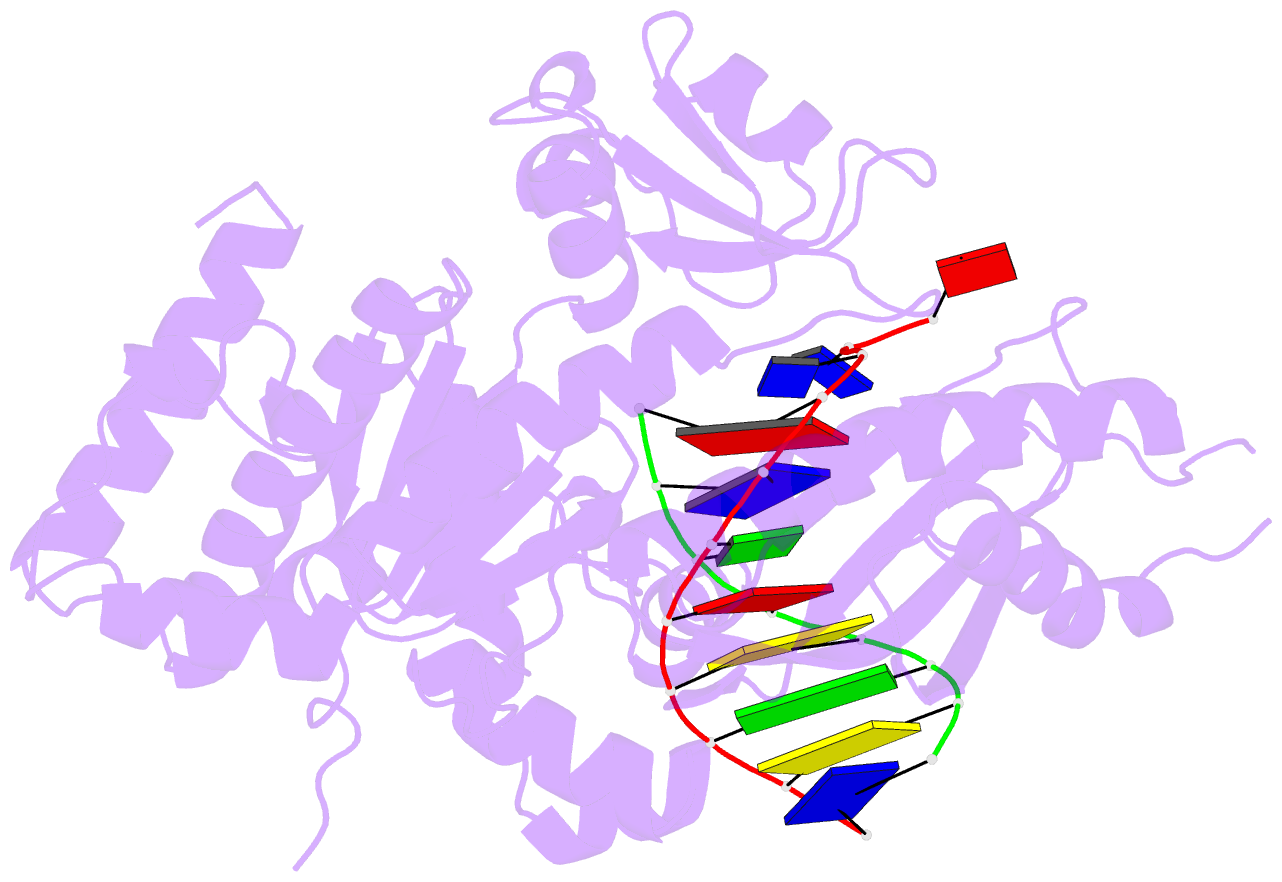
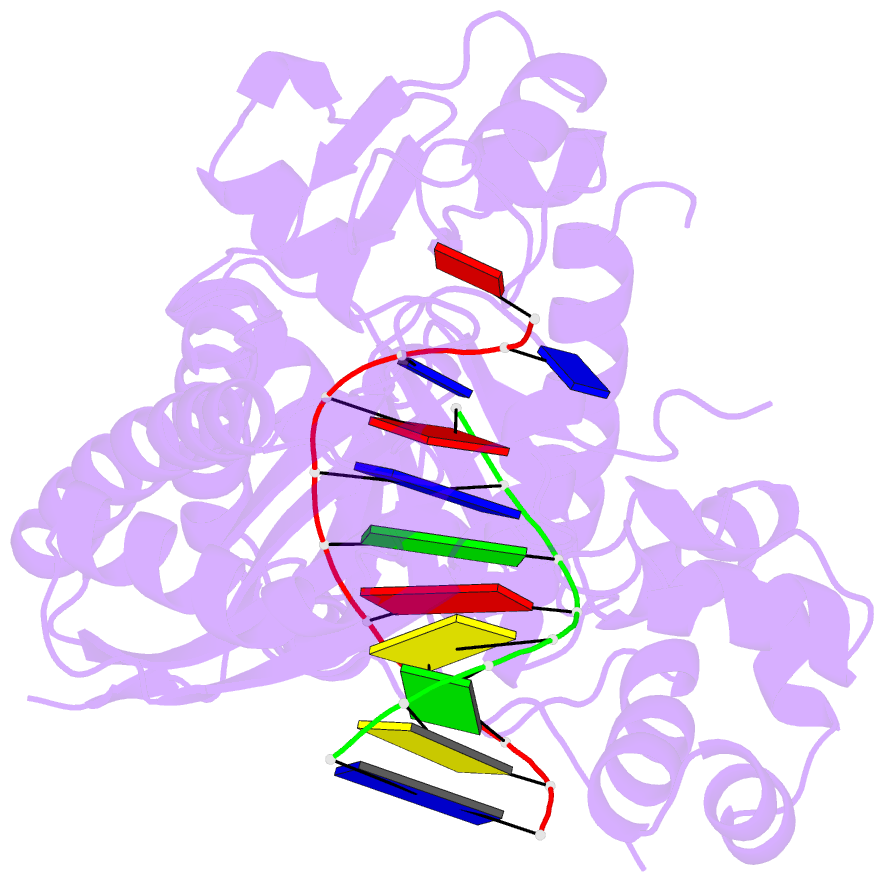
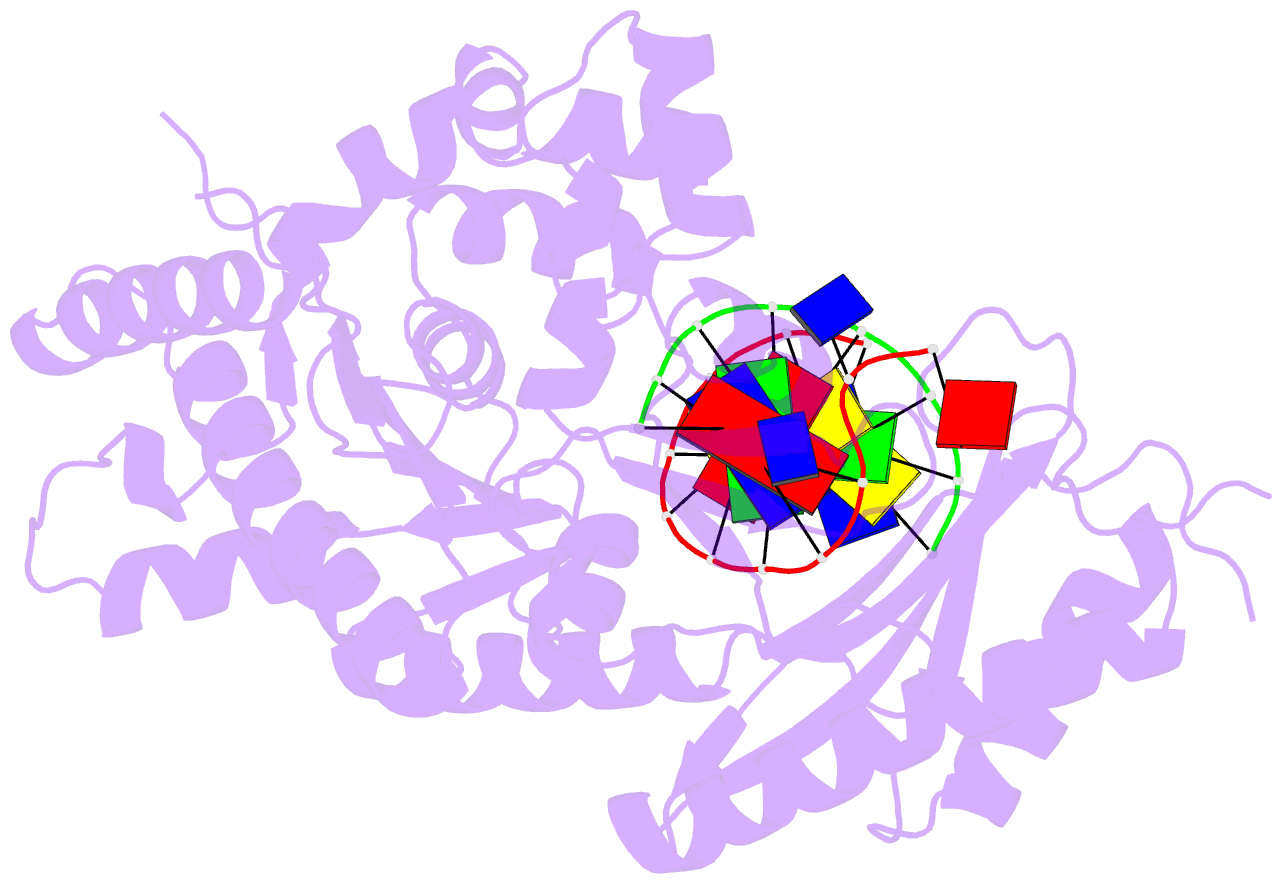
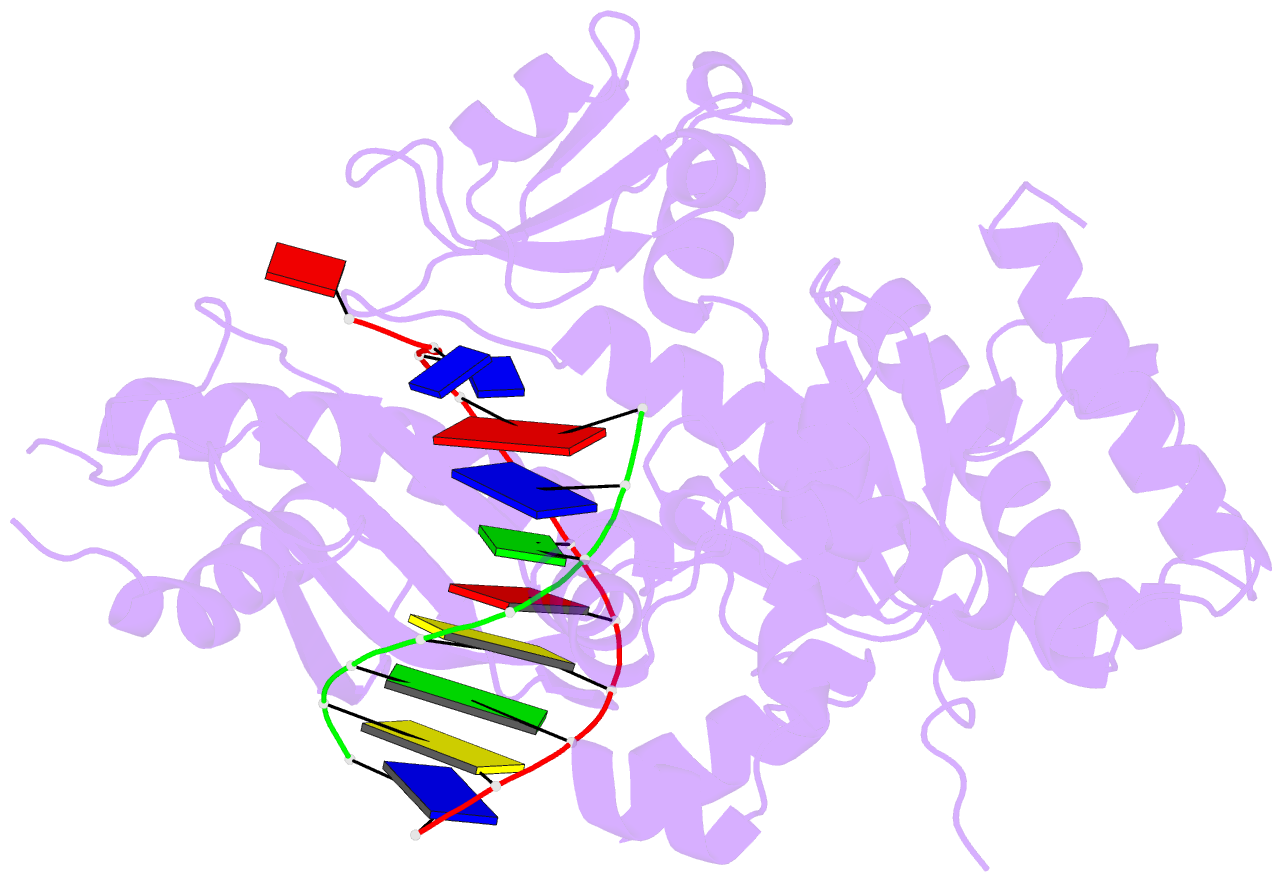
- Human DNA polymerase eta in binary complex with DNA
- Reference
- Ummat A, Silverstein TD, Jain R, Buku A, Johnson RE, Prakash L, Prakash S, Aggarwal AK (2012): "Human DNA Polymerase Eta Is Pre-Aligned for dNTP Binding and Catalysis." J.Mol.Biol., 415, 627-634. doi: 10.1016/j.jmb.2011.11.038.
- Abstract
- Pre-steady-state kinetic studies on Y-family DNA polymerase η (Polη) have suggested that the polymerase undergoes a rate-limiting conformational change step before the phosphoryl transfer of the incoming nucleotide to the primer terminus. However, the nature of this rate-limiting conformational change step has been unclear, due in part to the lack of structural information on the Polη binary complex. We present here for the first time a crystal structure of human Polη (hPolη) in binary complex with its DNA substrate. We show that the hPolη domains move only slightly on dNTP binding and that the polymerase by and large is pre-aligned for dNTP binding and catalysis. We also show that there is no major reorientation of the DNA from a nonproductive to a productive configuration and that the active site is devoid of metals in the absence of dNTP. Together, these observations lead us to suggest that the rate-limiting conformational change step in the Polη replication cycle likely corresponds to a rate-limiting entry of catalytic metals in the active site.