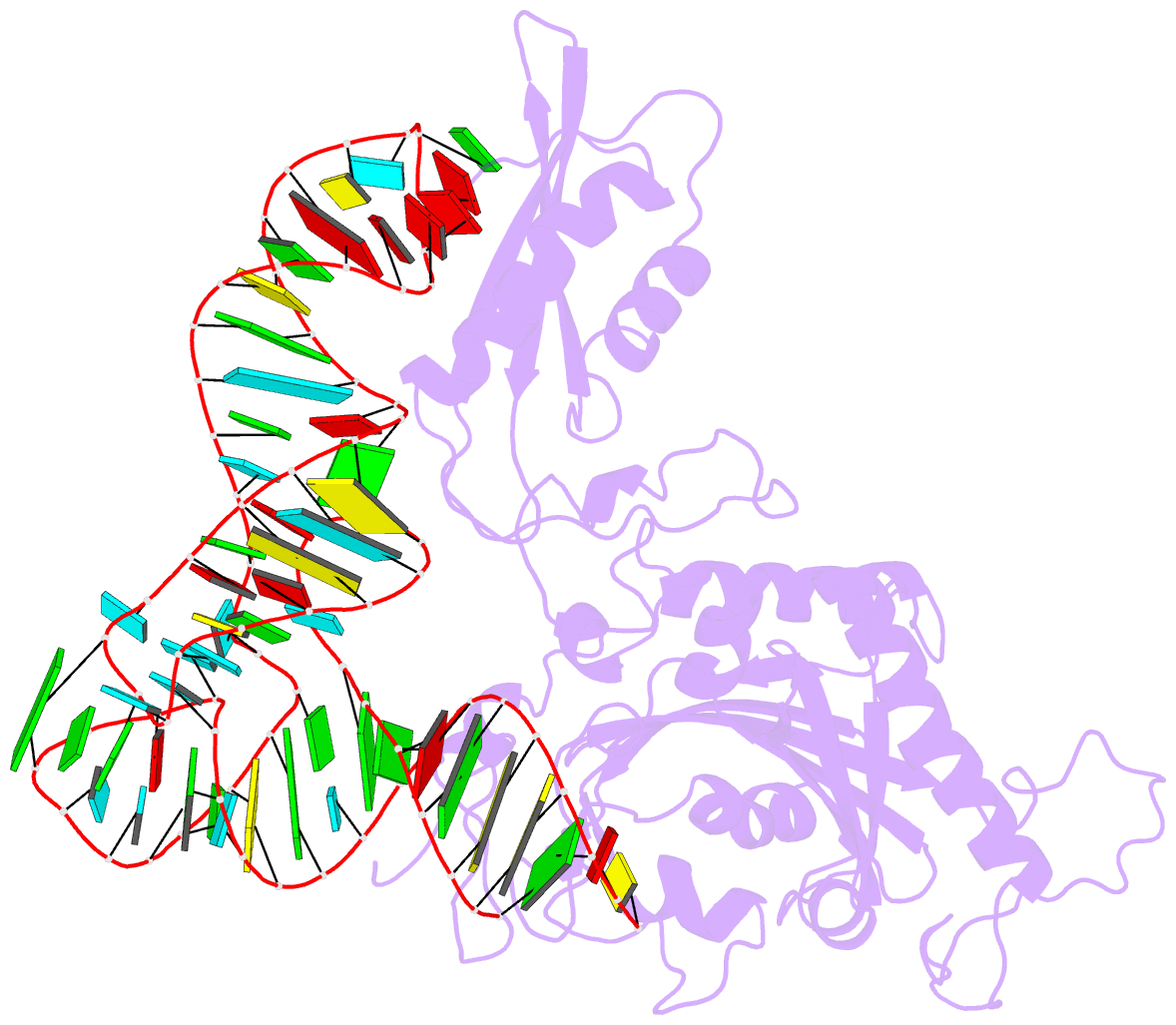
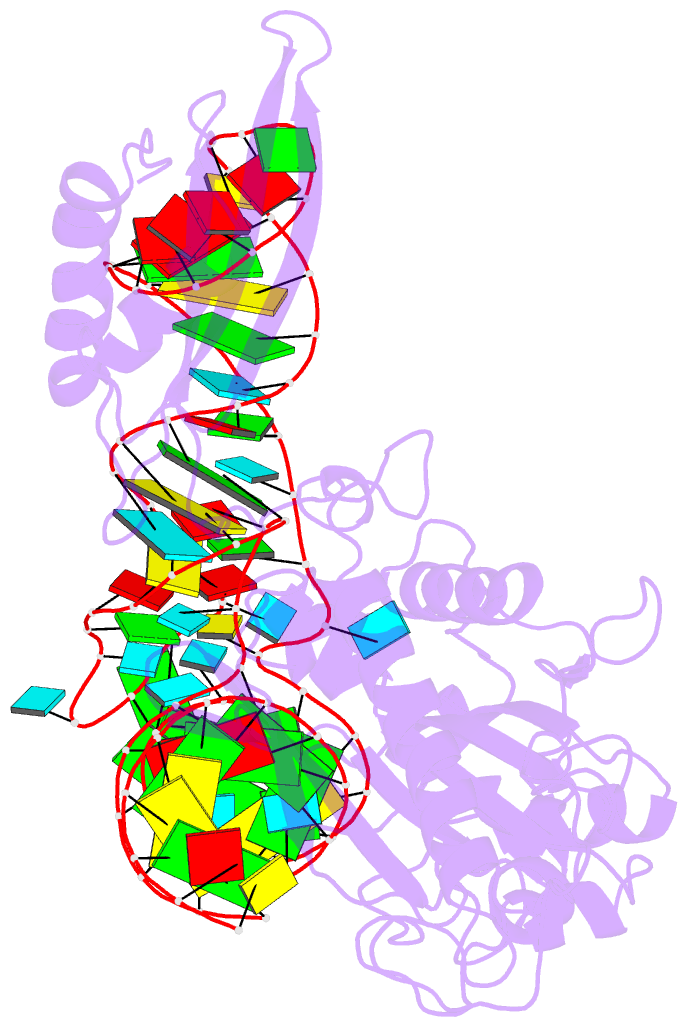
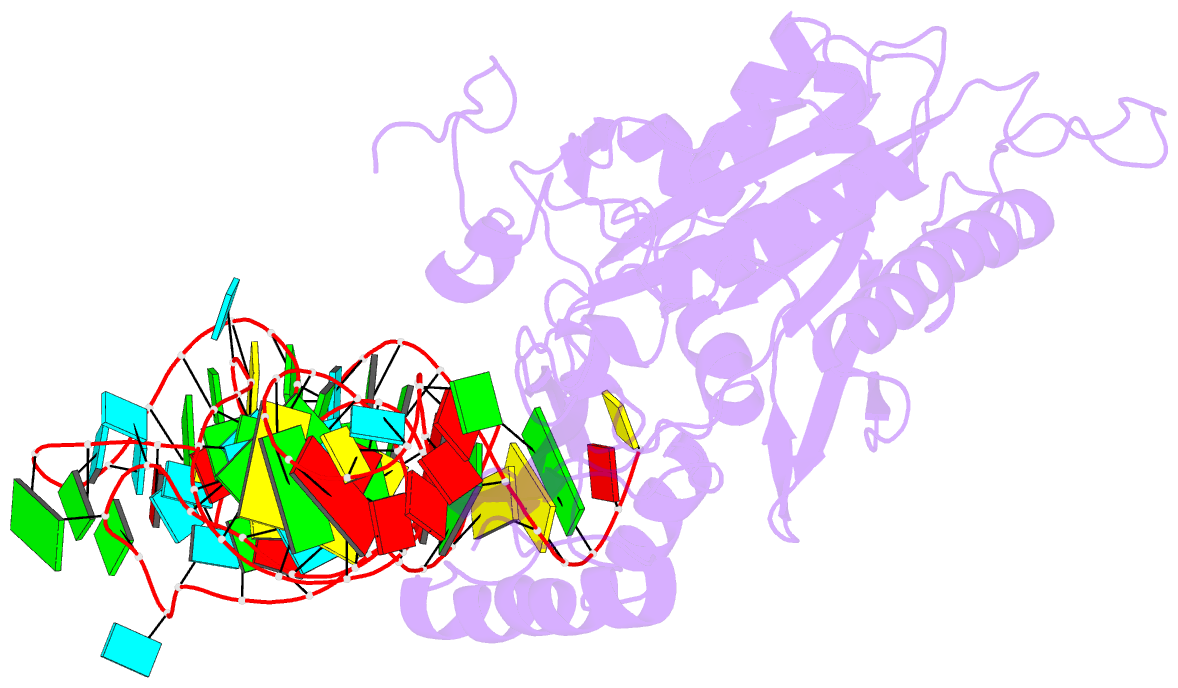
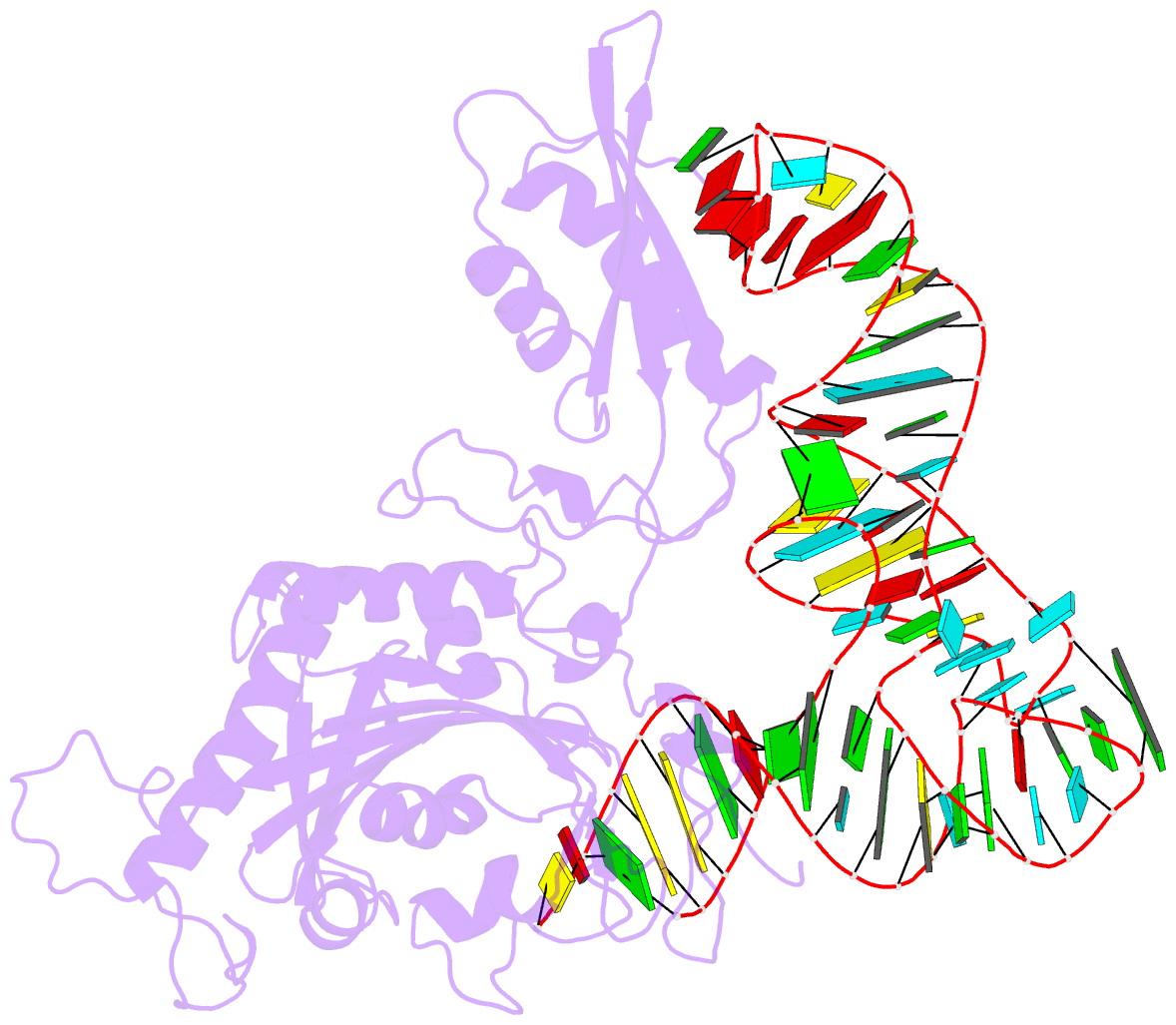
Summary information and primary citation
- PDB-id
- 3tup; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (3.05 Å)
- Summary
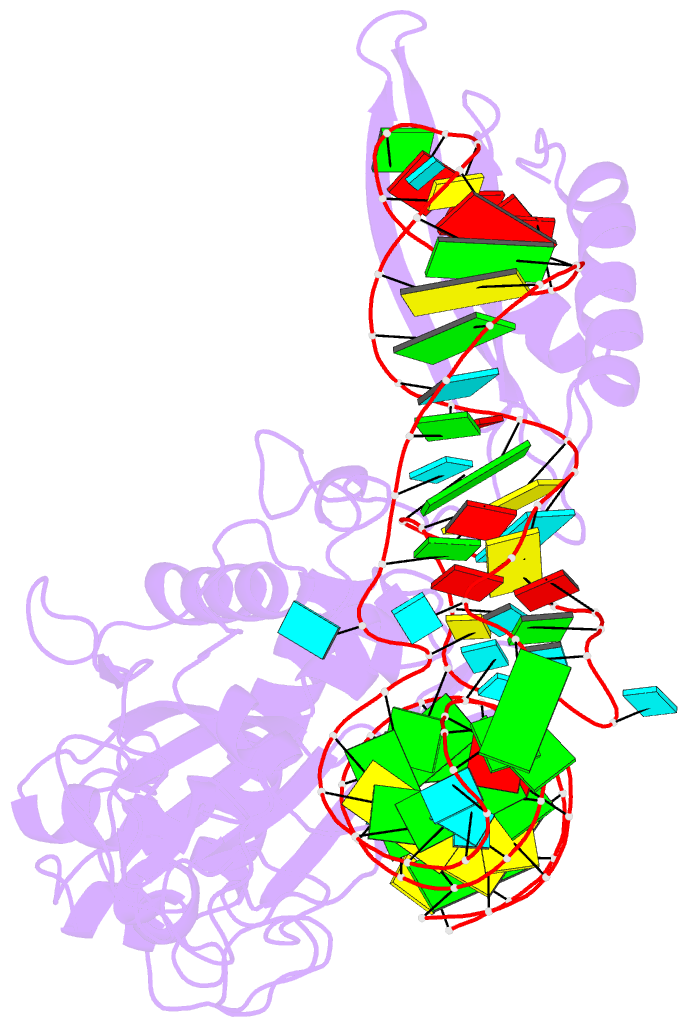
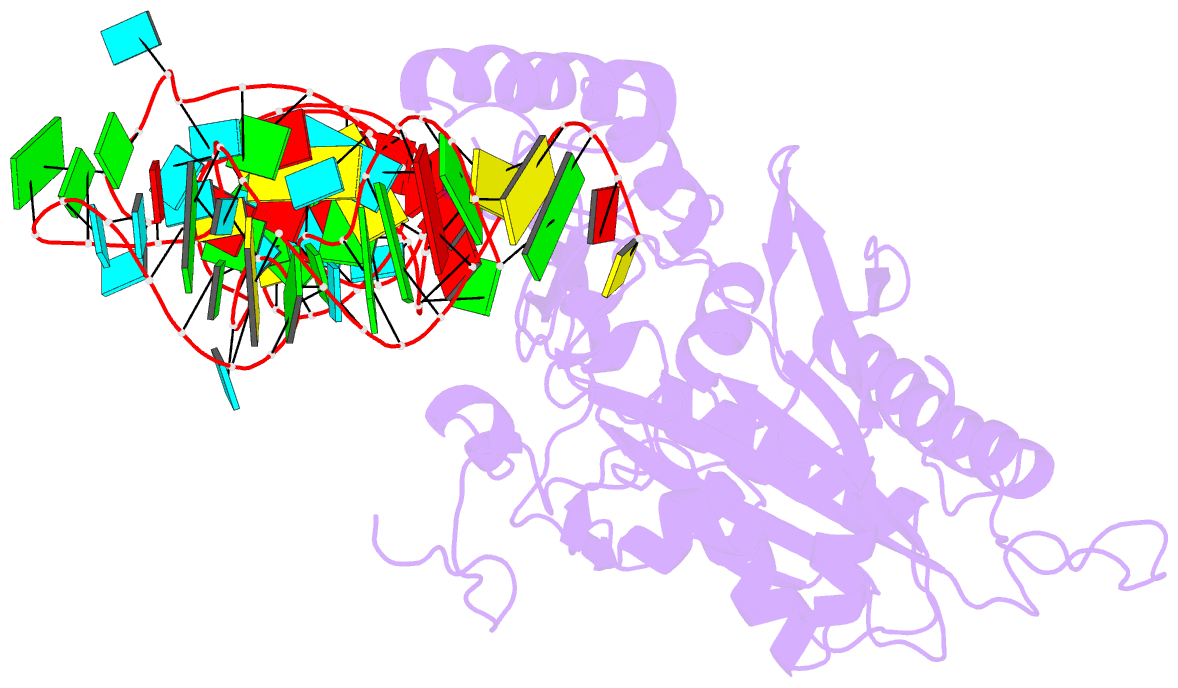
- Crystal structure of human mitochondrial phers complexed with trnaphe in the active open state
- Reference
- Klipcan L, Moor N, Finarov I, Kessler N, Sukhanova M, Safro MG (2012): "Crystal Structure of Human Mitochondrial PheRS Complexed with tRNA(Phe) in the Active "Open" State." J.Mol.Biol., 415, 527-537. doi: 10.1016/j.jmb.2011.11.029.
- Abstract
- Monomeric human mitochondrial phenylalanyl-tRNA synthetase (PheRS), or hmPheRS, is the smallest known enzyme exhibiting aminoacylation activity. HmPheRS consists of only two structural domains and differs markedly from heterodimeric eukaryotic cytosolic and bacterial analogs both in the domain organization and in the mode of tRNA binding. Here, we describe the first crystal structure of mitochondrial aminoacyl-tRNA synthetase (aaRS) complexed with tRNA at a resolution of 3.0 Å. Unlike bacterial PheRSs, the hmPheRS recognizes C74, the G1-C72 base pair, and the "discriminator" base A73, proposed to contribute to tRNA(Phe) identity in the yeast mitochondrial enzyme. An interaction of the tRNA acceptor stem with the signature motif 2 residues of hmPheRS is of critical importance for the stabilization of the CCA-extended conformation and its correct placement in the synthetic site of the enzyme. The crystal structure of hmPheRS-tRNA(Phe) provides direct evidence that the formation of the complex with tRNA requires a significant rearrangement of the anticodon-binding domain from the "closed" to the productive "open" state. Global repositioning of the domain is tRNA modulated and governed by long-range electrostatic interactions.