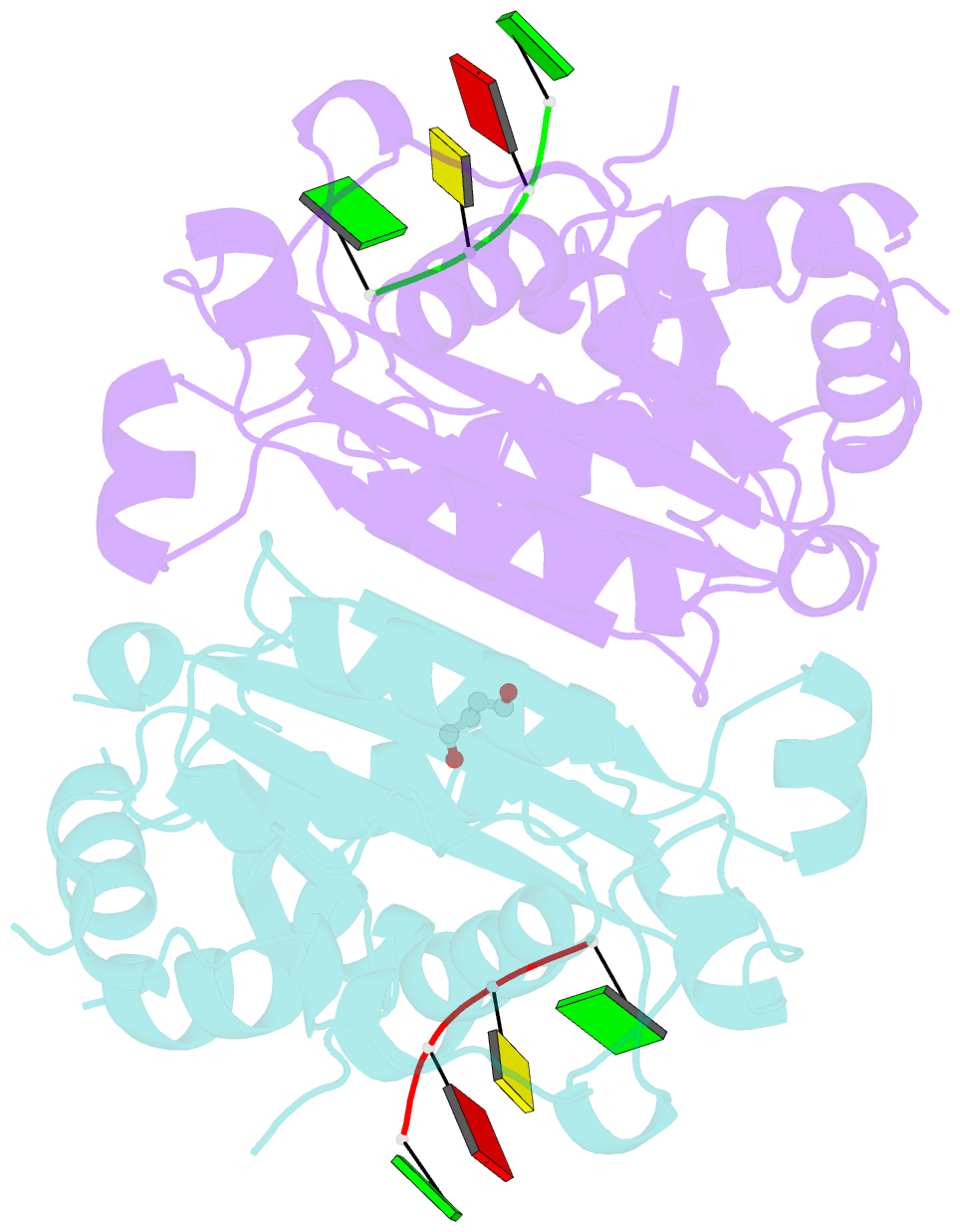
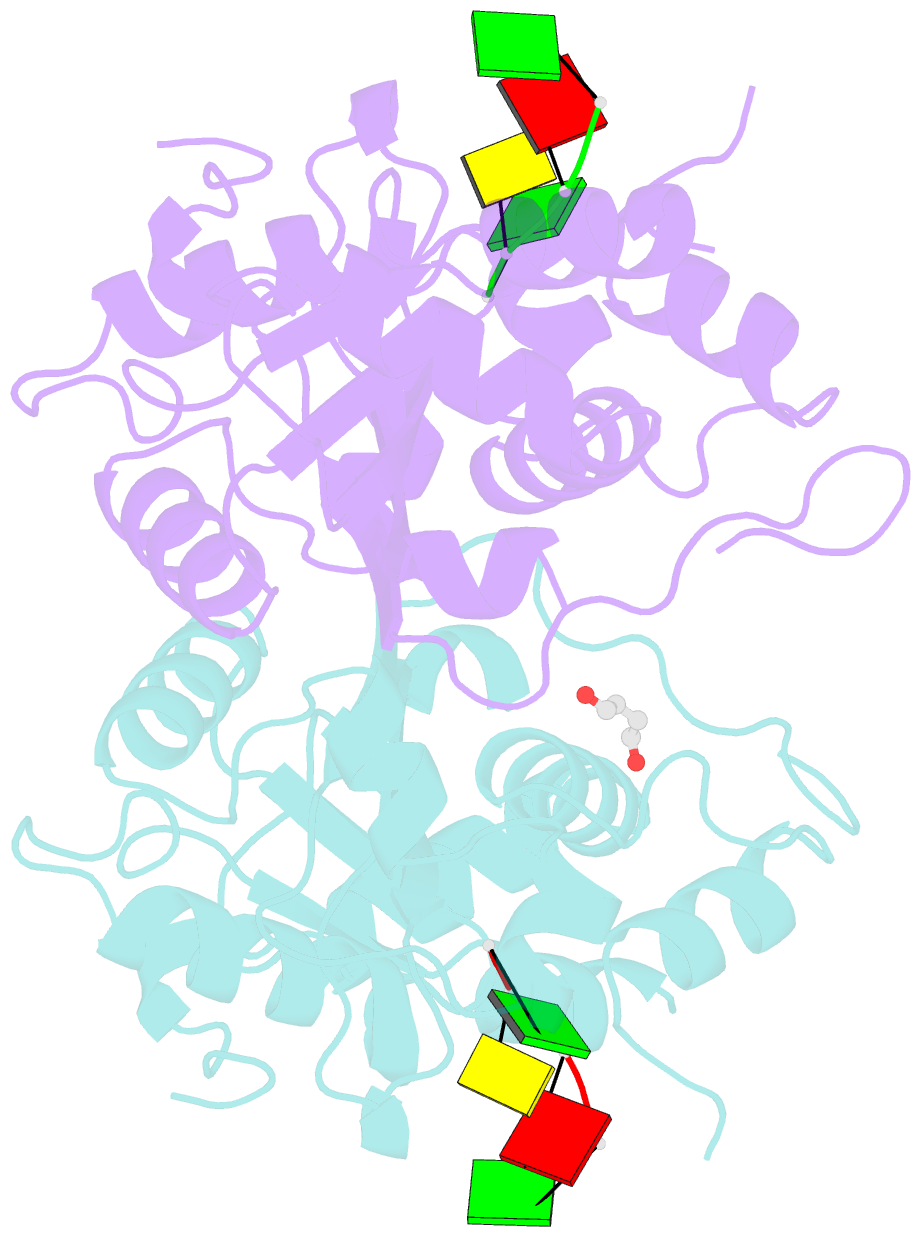
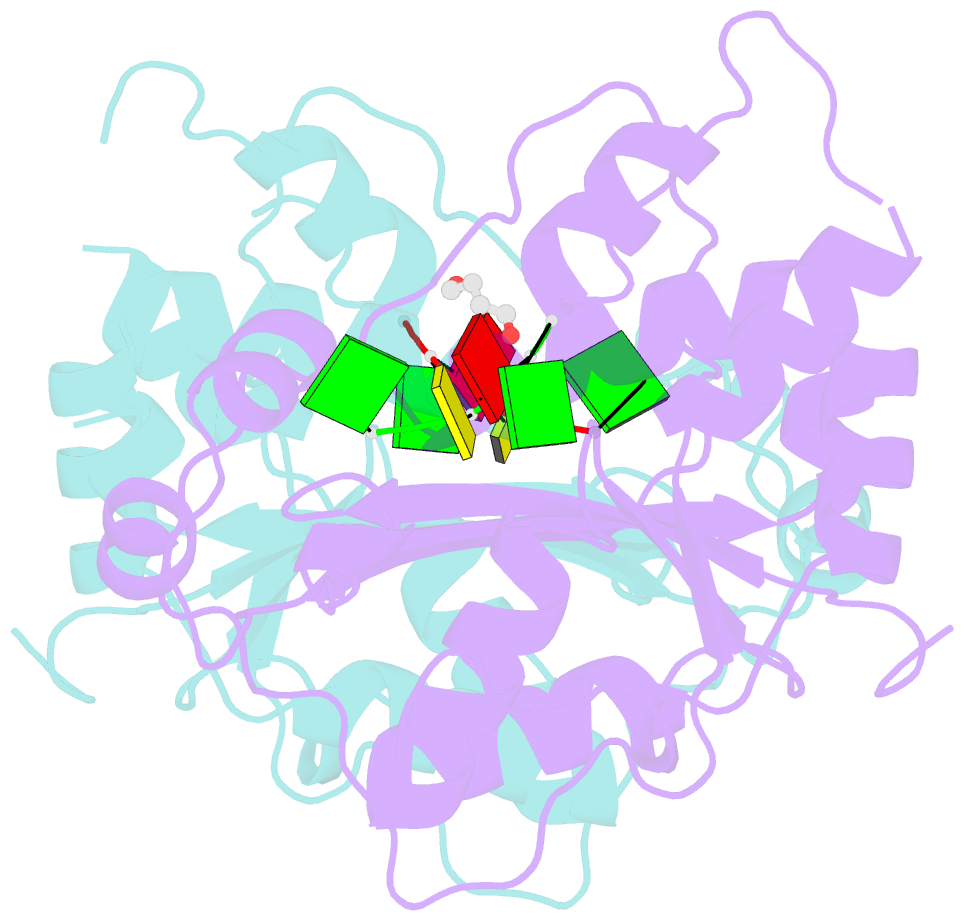
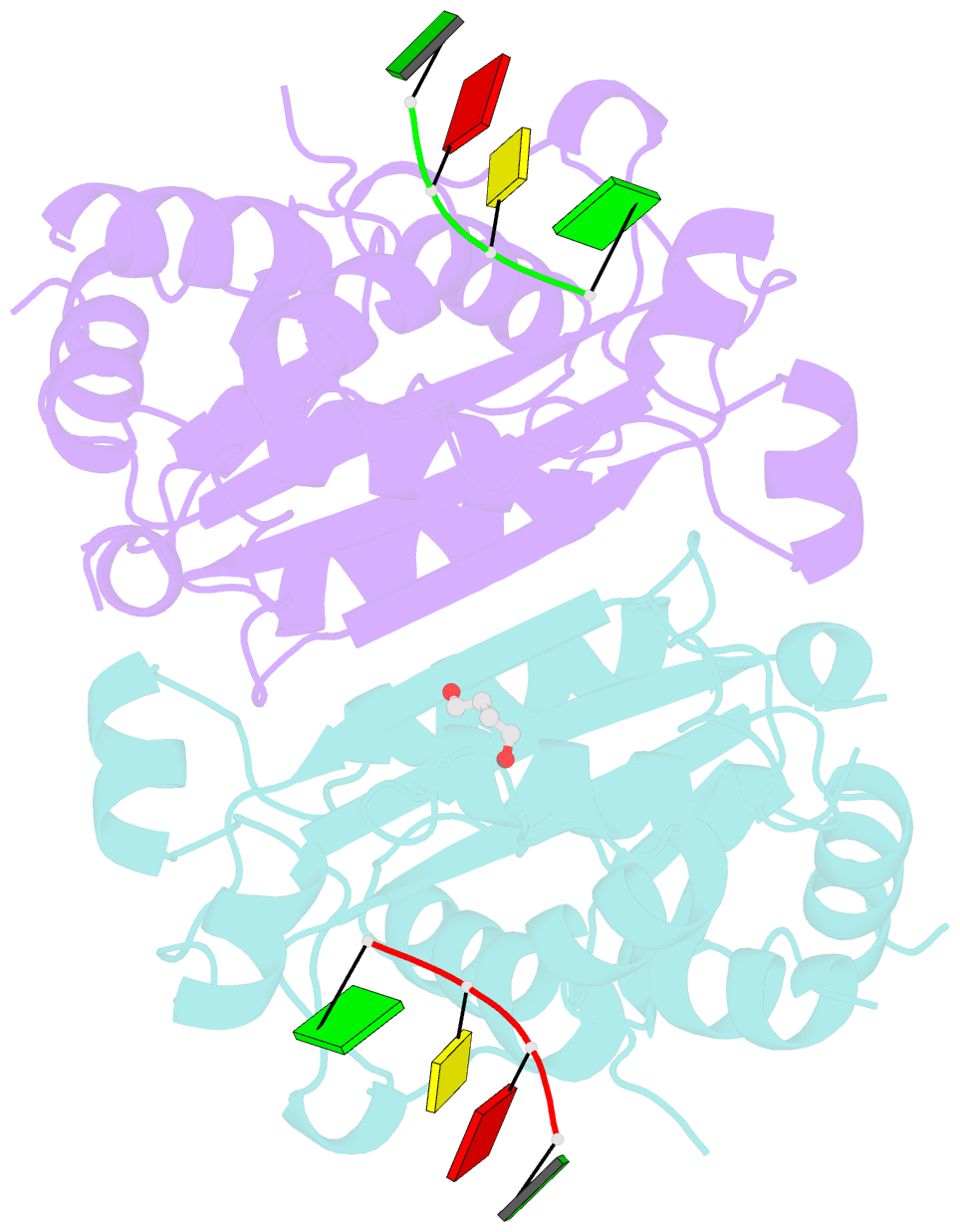
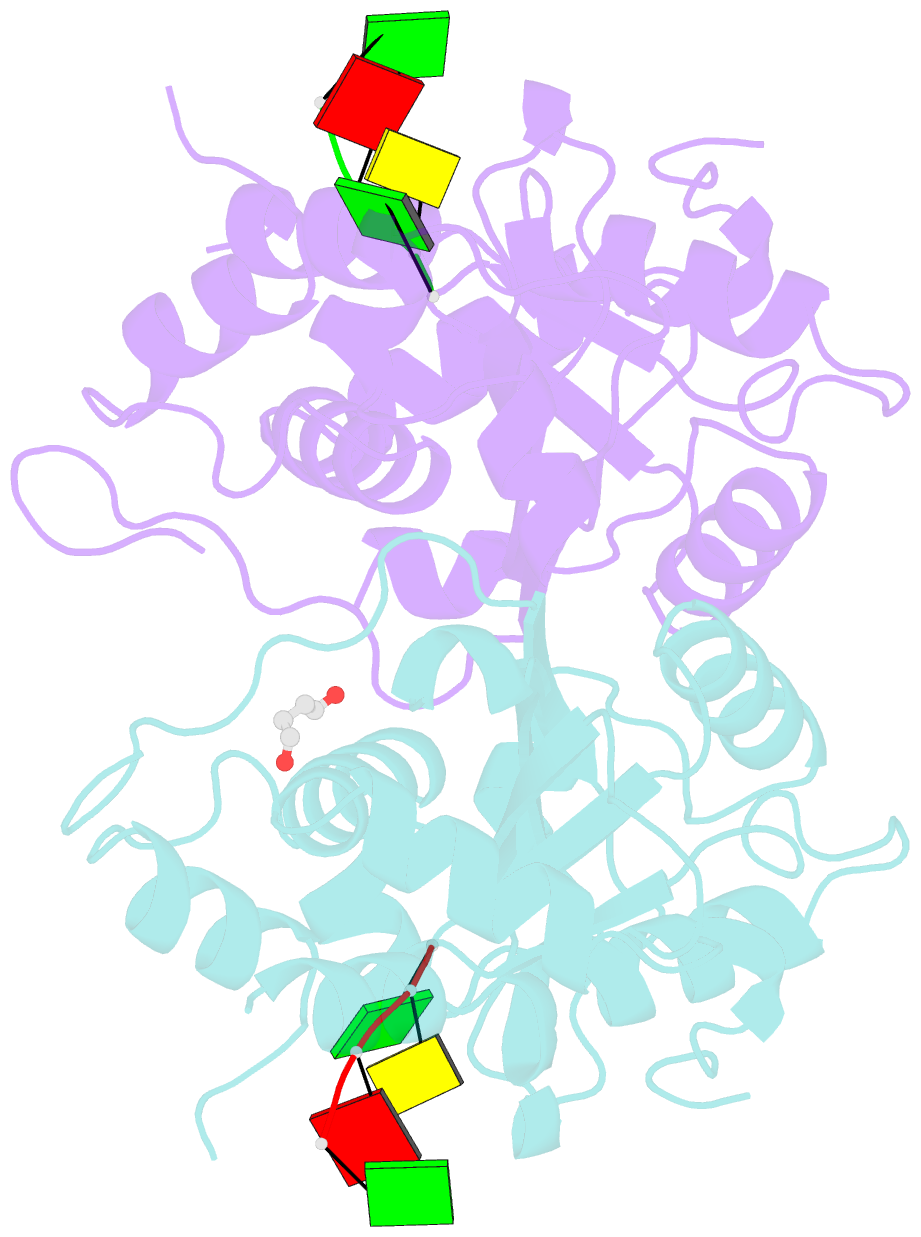
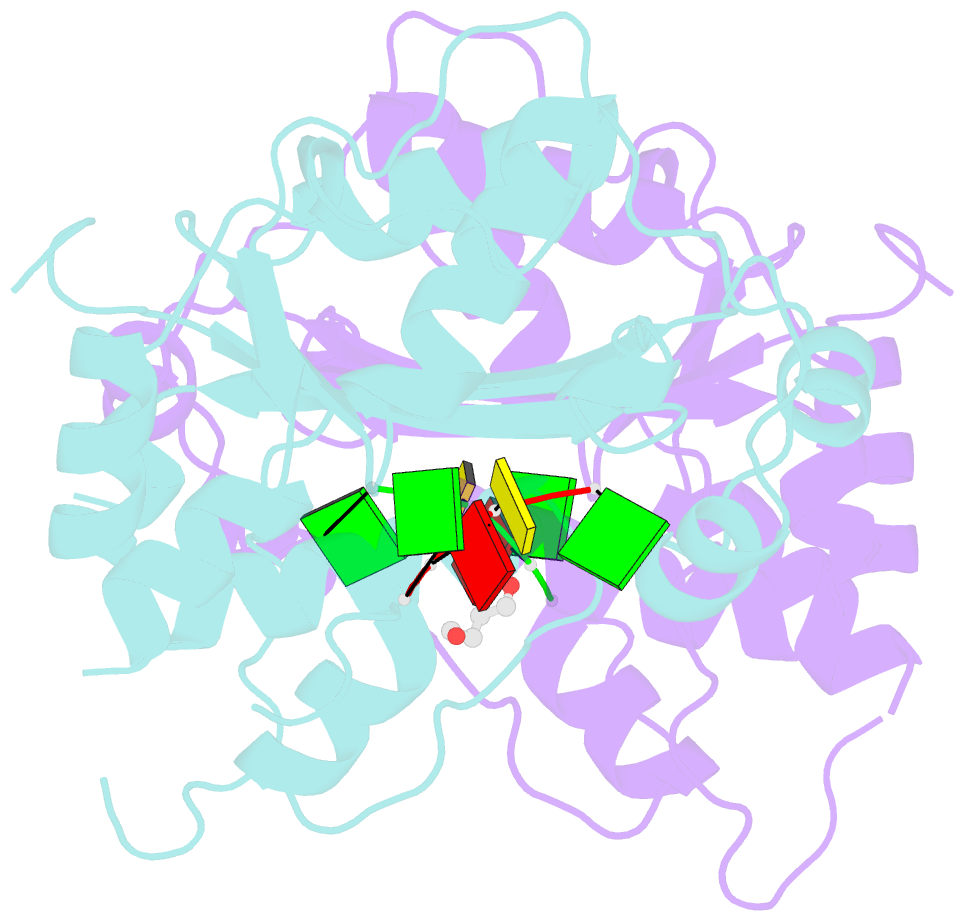
Summary information and primary citation
- PDB-id
- 3u3y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.28 Å)
- Summary
- Mouse trex1 d200h mutant
- Reference
- Bailey SL, Harvey S, Perrino FW, Hollis T (2012): "Defects in DNA degradation revealed in crystal structures of TREX1 exonuclease mutations linked to autoimmune disease." Dna Repair, 11, 65-73. doi: 10.1016/j.dnarep.2011.10.007.
- Abstract
- Mutations within the human TREX1 3' exonuclease are associated with Aicardi-Goutières Syndrome (AGS) and familial chilblain lupus (FCL). Both AGS and FCL are autoimmune diseases that result in increased levels of interferon alpha and circulating antibodies to DNA. TREX1 is a member of the endoplasmic reticulum (ER)-associated SET complex and participates in granzyme A-mediated cell death to degrade nicked genomic DNA. The loss of TREX1 activity may result in the accumulation of double-stranded DNA (dsDNA) degradation intermediates that trigger autoimmune activation. The X-ray crystal structures of the TREX1 wt apoprotein, the dominant D200H, D200N and D18N homodimer mutants derived from AGS and FCL patients, as well as the recessive V201D homodimer mutant have been determined. The structures of the D200H and D200N mutant proteins reveal the enzyme has lost coordination of one of the active site metals, and the catalytic histidine (H195) is trapped in a conformation pointing away from the active site. The TREX1 D18N and V201D mutants are able to bind both metals in the active site, but with inter-metal distances that are larger than optimal for catalysis. Additionally, all of the mutant structures reveal a reduced mobility in the catalytic histidine, providing further explanation for the loss of catalytic activity. The structures of the mutant TREX1 proteins provide insight into the dysfunction relating to human disease. Additionally, the TREX1 apoprotein structure together with the previously determined wild type substrate and product structures allow us to propose a distinct mechanism for the TREX1 exonuclease.