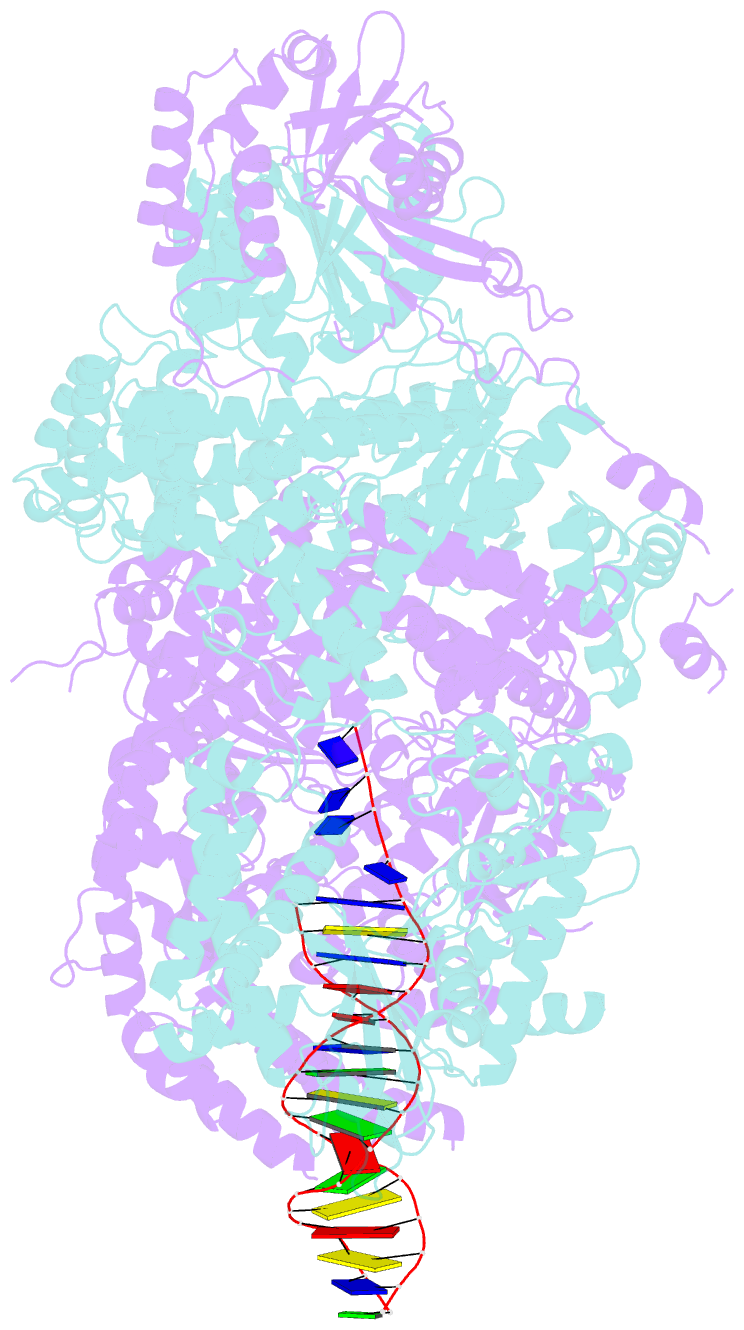
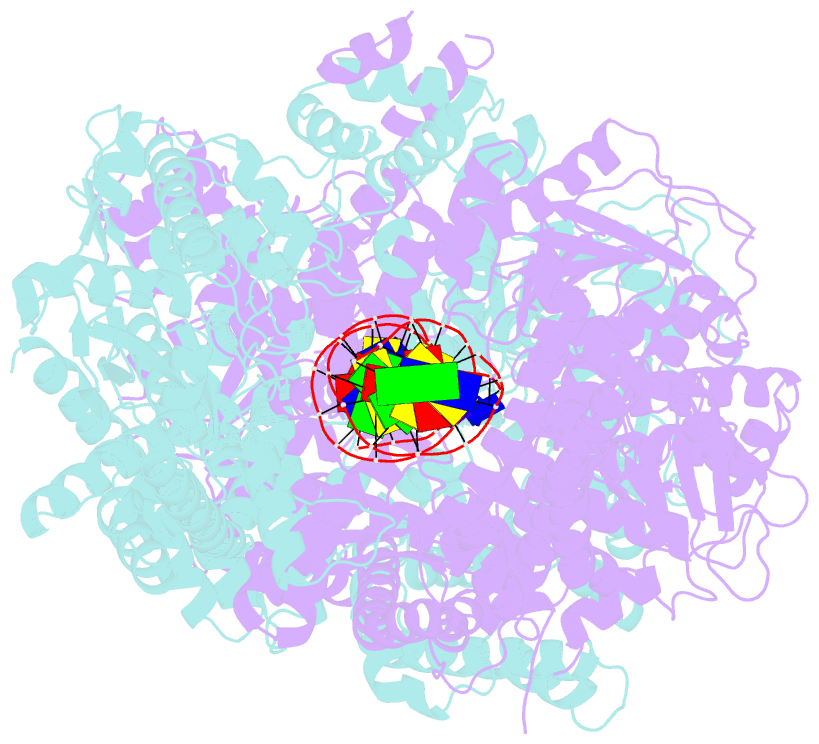
Summary information and primary citation
- PDB-id
- 3u44; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.201 Å)
- Summary
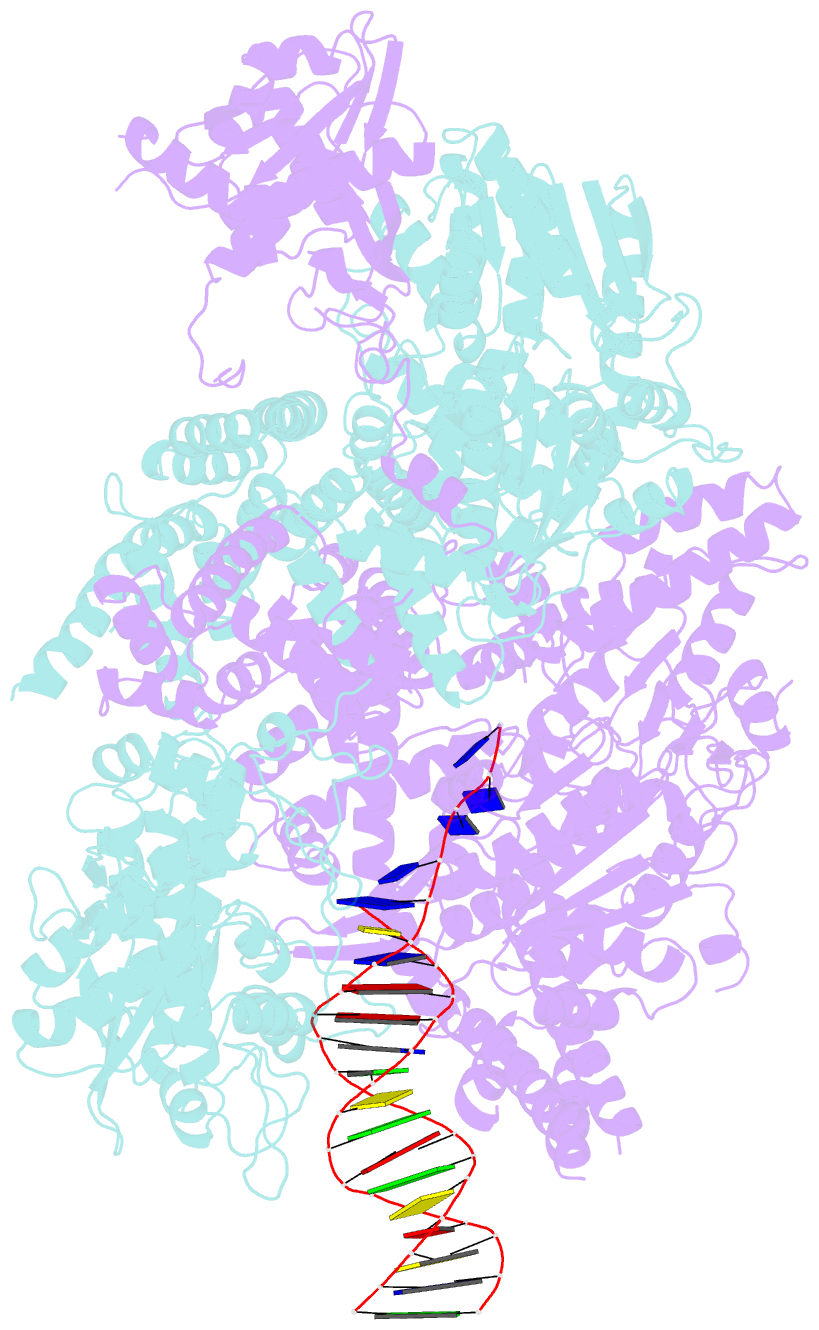
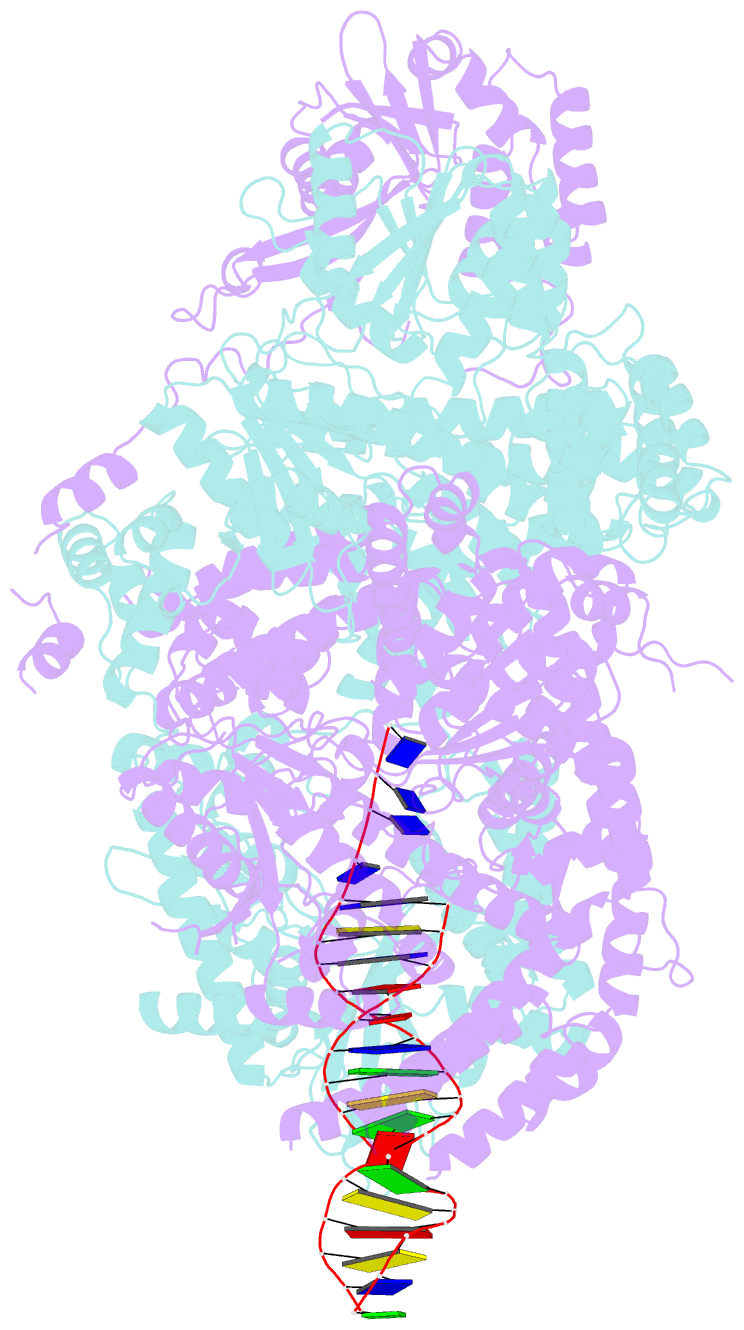
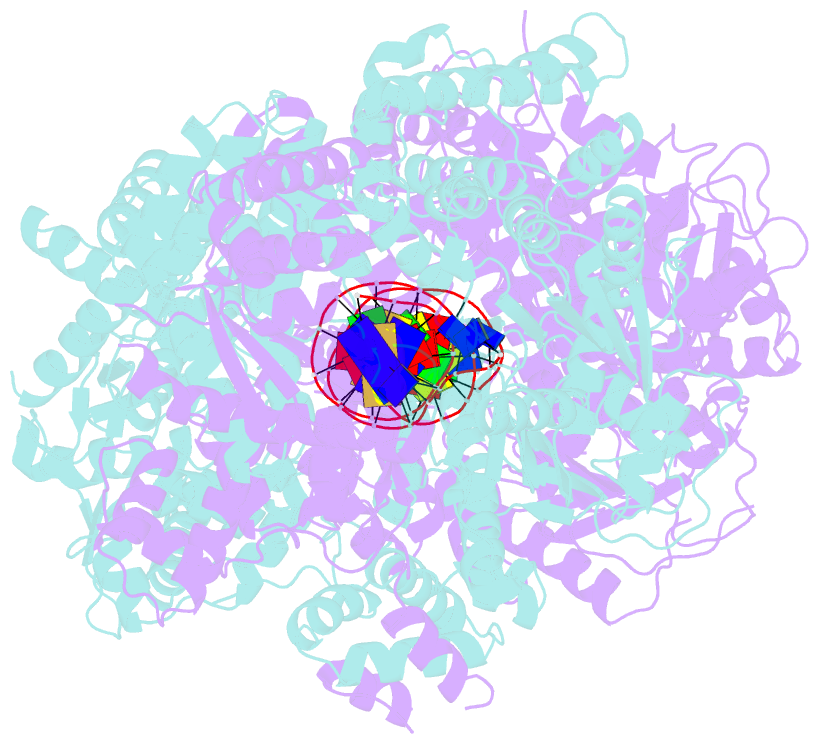
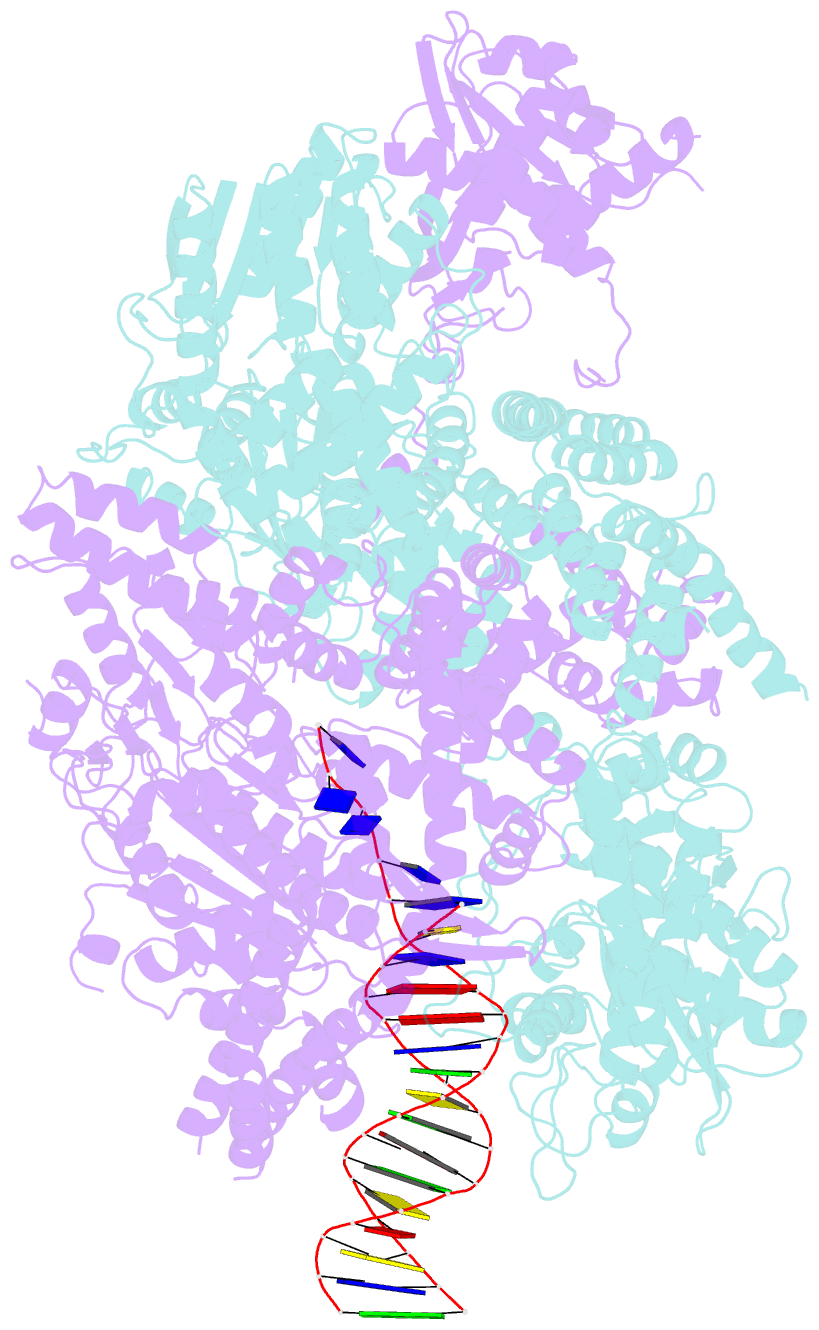
- Crystal structure of addab-DNA complex
- Reference
- Saikrishnan K, Yeeles JT, Gilhooly NS, Krajewski WW, Dillingham MS, Wigley DB (2012): "Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex." Embo J., 31, 1568-1578. doi: 10.1038/emboj.2012.9.
- Abstract
- In bacterial cells, processing of double-stranded DNA breaks for repair by homologous recombination is dependent upon the recombination hotspot sequence Chi and is catalysed by either an AddAB- or RecBCD-type helicase-nuclease. Here, we report the crystal structure of AddAB bound to DNA. The structure allows identification of a putative Chi-recognition site in an inactivated helicase domain of the AddB subunit. By generating mutant protein complexes that do not respond to Chi, we show that residues responsible for Chi recognition are located in positions equivalent to the signature motifs of a conventional helicase. Comparison with the related RecBCD complex, which recognizes a different Chi sequence, provides further insight into the structural basis for sequence-specific ssDNA recognition. The structure suggests a simple mechanism for DNA break processing, explains how AddAB and RecBCD can accomplish the same overall reaction with different sets of functional modules and reveals details of the role of an Fe-S cluster in protein stability and DNA binding.