Summary information and primary citation
- PDB-id
- 3u61; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.2 Å)
- Summary
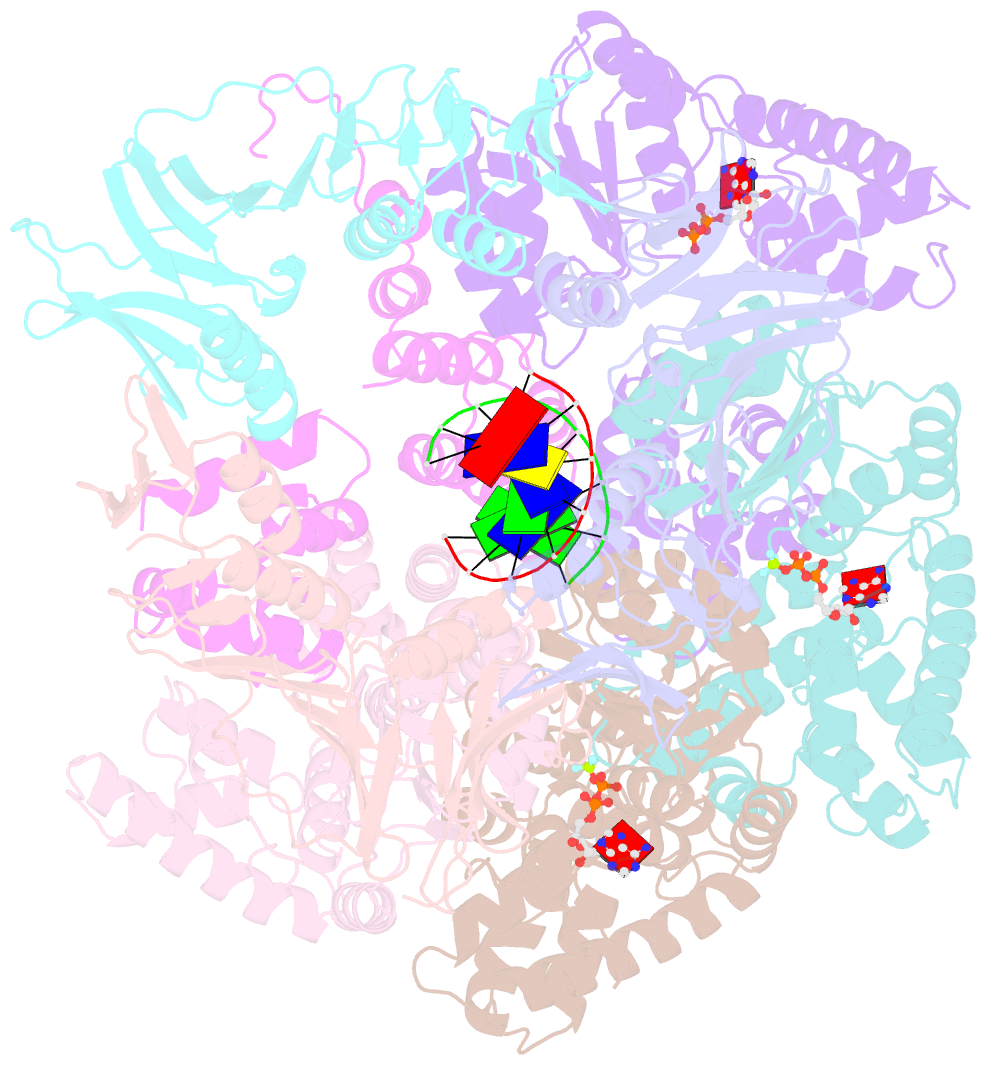
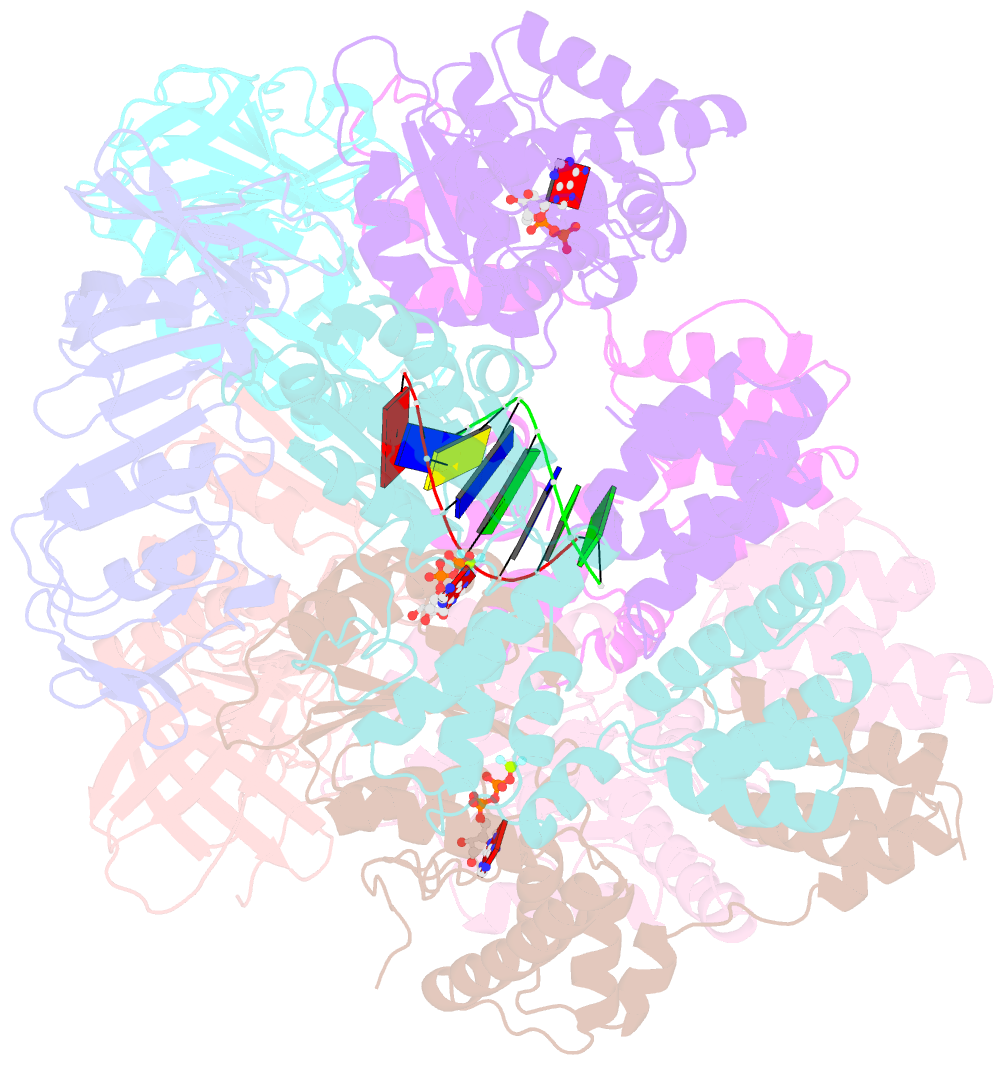
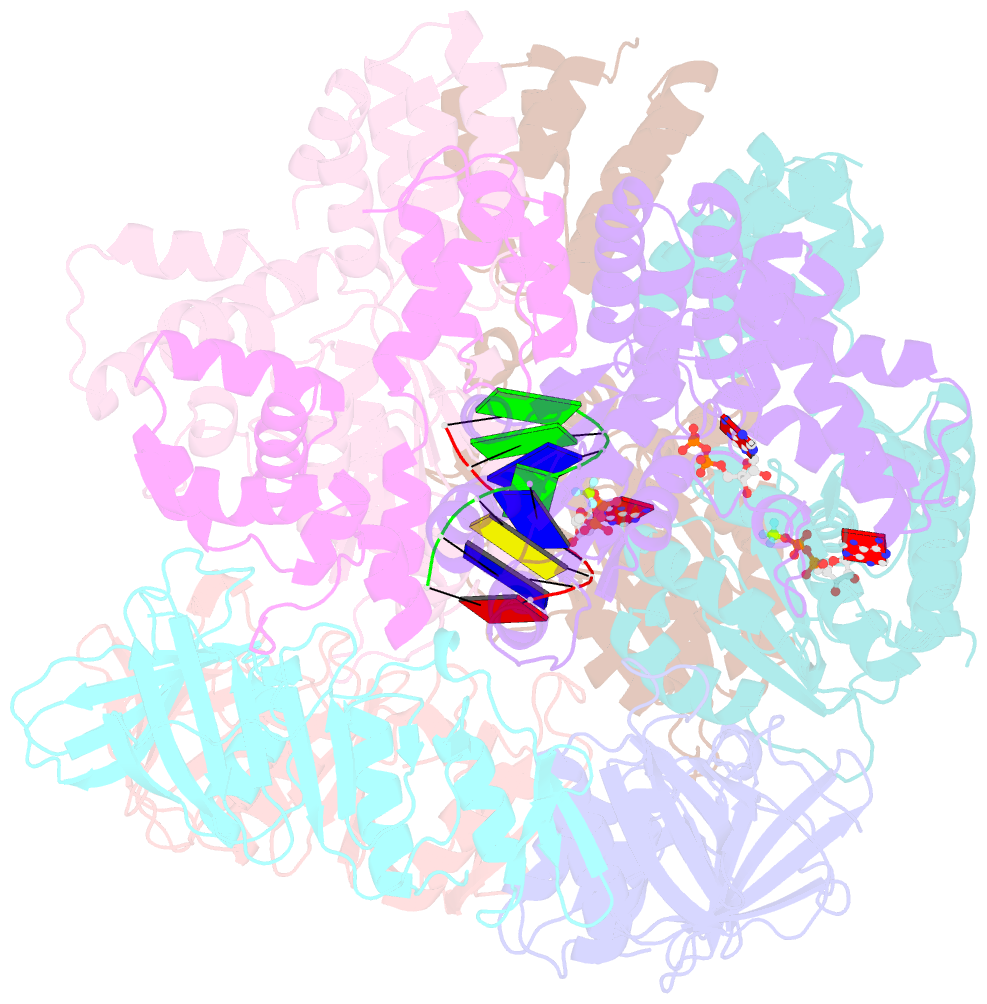
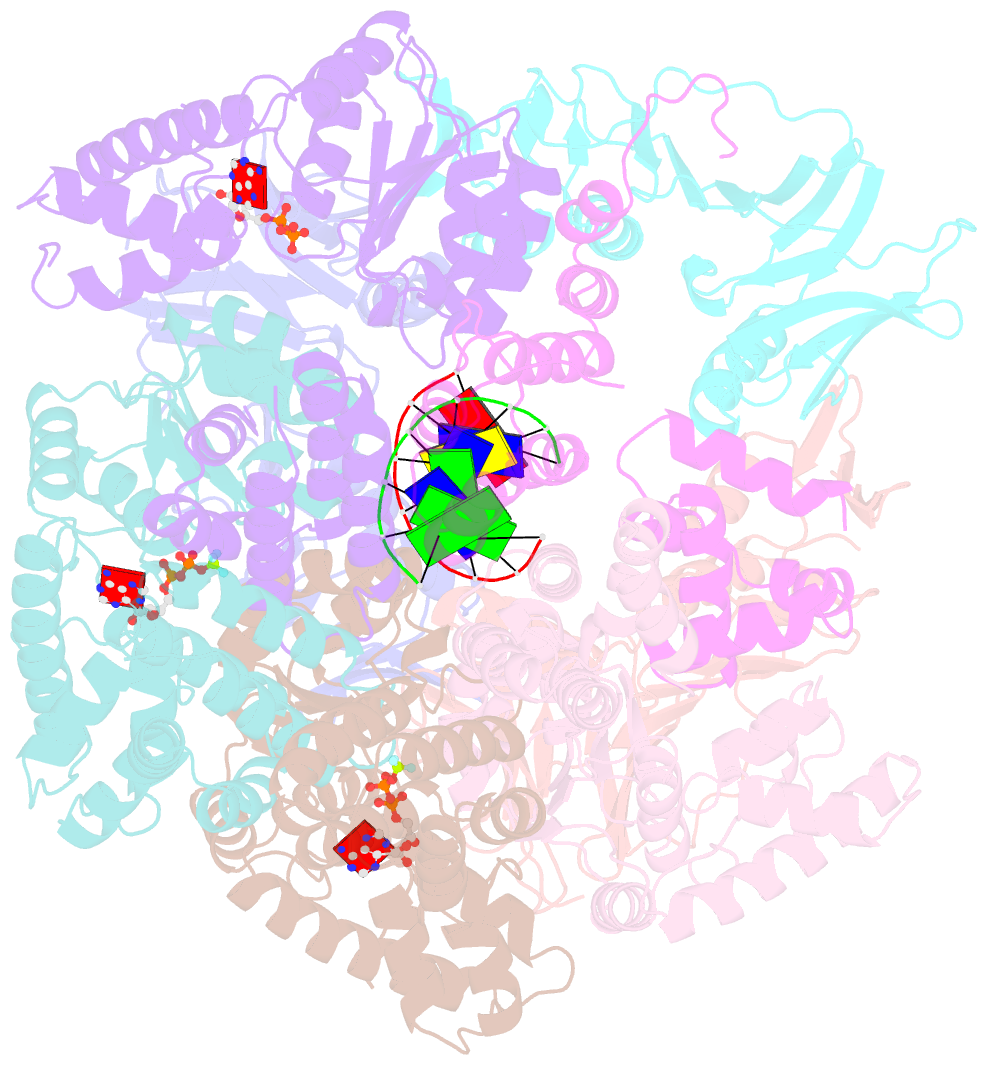
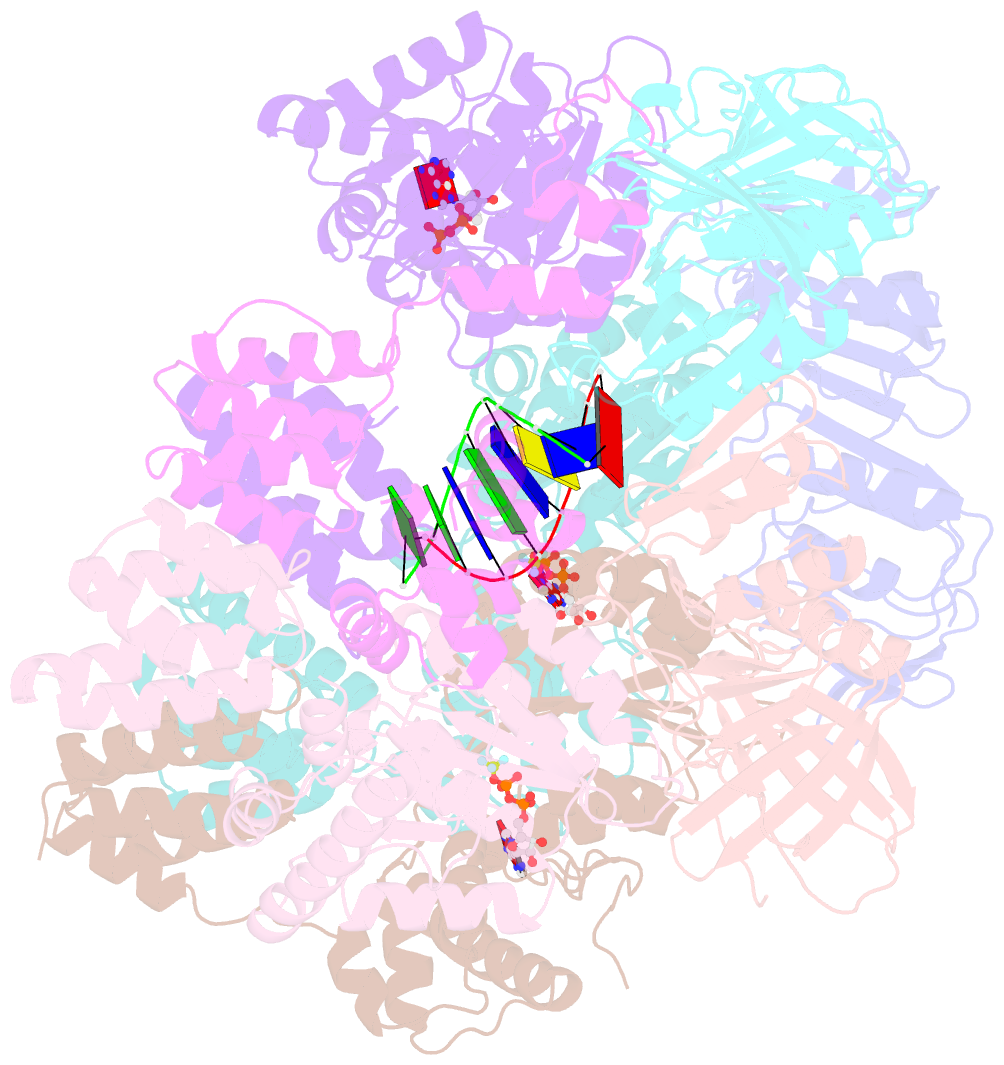
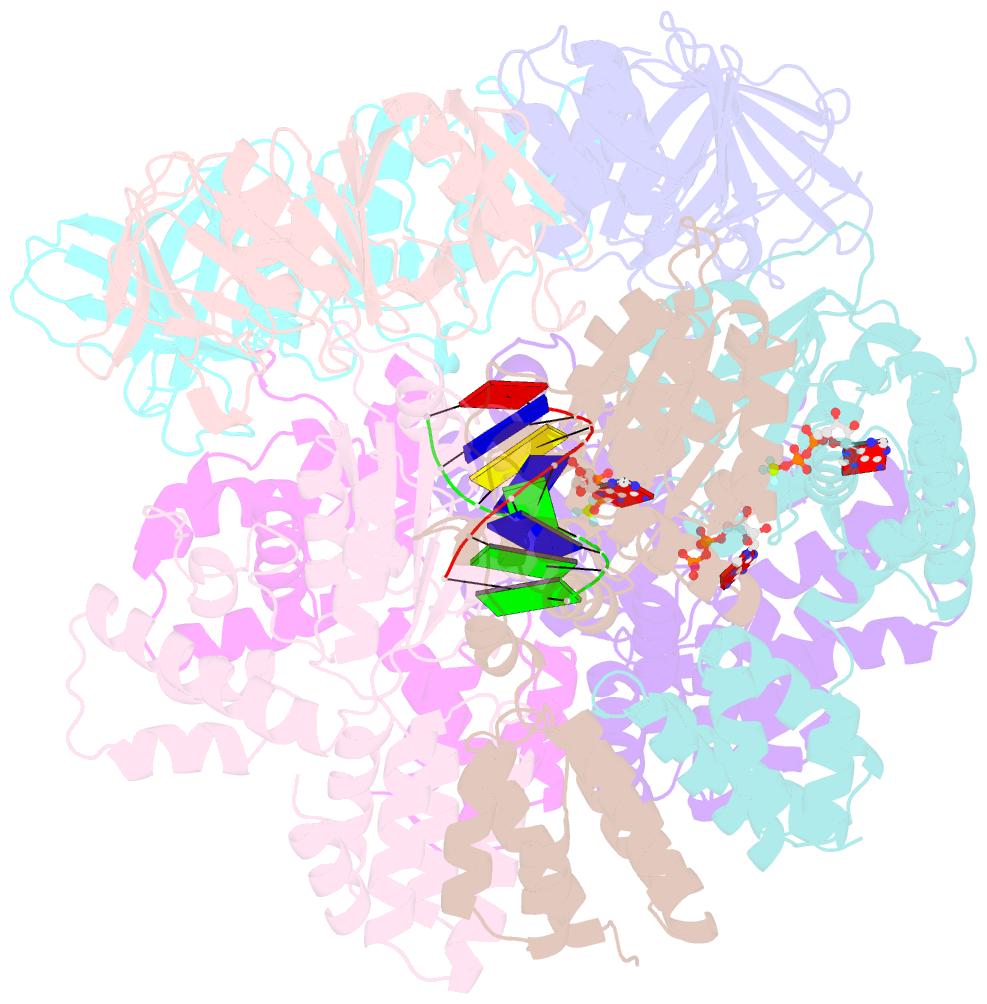
- Structure of t4 bacteriophage clamp loader bound to closed clamp, DNA and atp analog and adp
- Reference
- Kelch BA, Makino DL, O'Donnell M, Kuriyan J (2011): "How a DNA polymerase clamp loader opens a sliding clamp." Science, 334, 1675-1680. doi: 10.1126/science.1211884.
- Abstract
- Processive chromosomal replication relies on sliding DNA clamps, which are loaded onto DNA by pentameric clamp loader complexes belonging to the AAA+ family of adenosine triphosphatases (ATPases). We present structures for the ATP-bound state of the clamp loader complex from bacteriophage T4, bound to an open clamp and primer-template DNA. The clamp loader traps a spiral conformation of the open clamp so that both the loader and the clamp match the helical symmetry of DNA. One structure reveals that ATP has been hydrolyzed in one subunit and suggests that clamp closure and ejection of the loader involves disruption of the ATP-dependent match in symmetry. The structures explain how synergy among the loader, the clamp, and DNA can trigger ATP hydrolysis and release of the closed clamp on DNA.