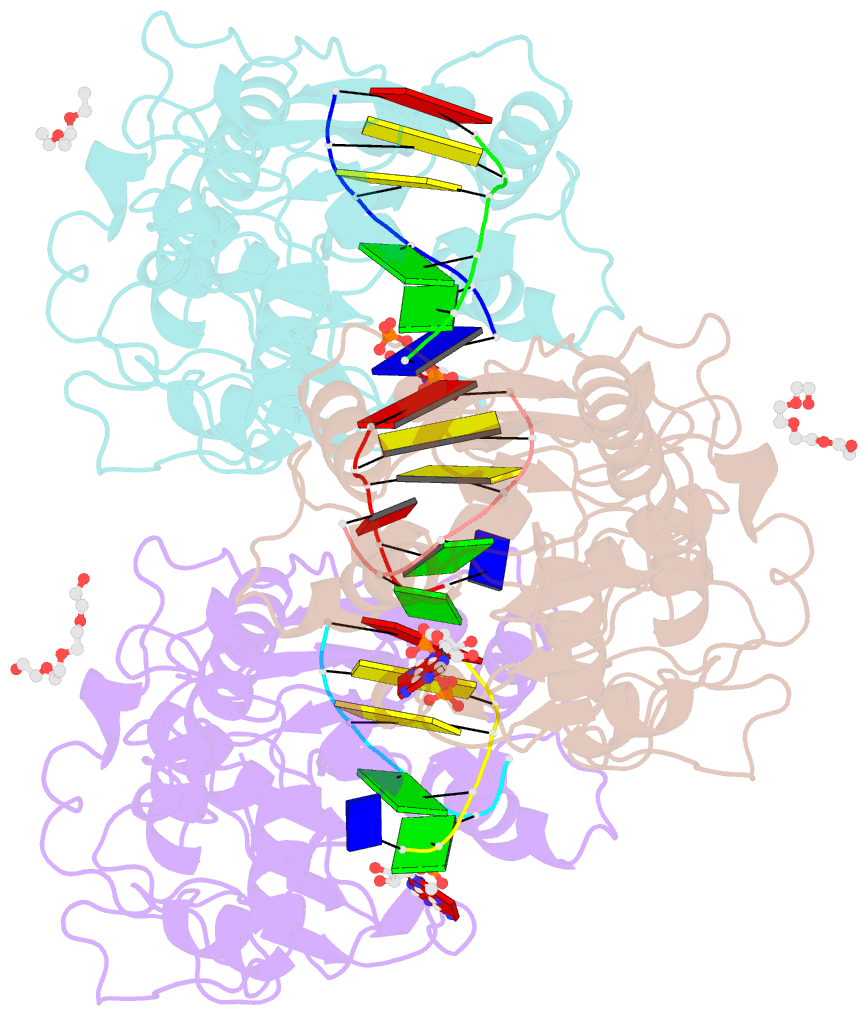
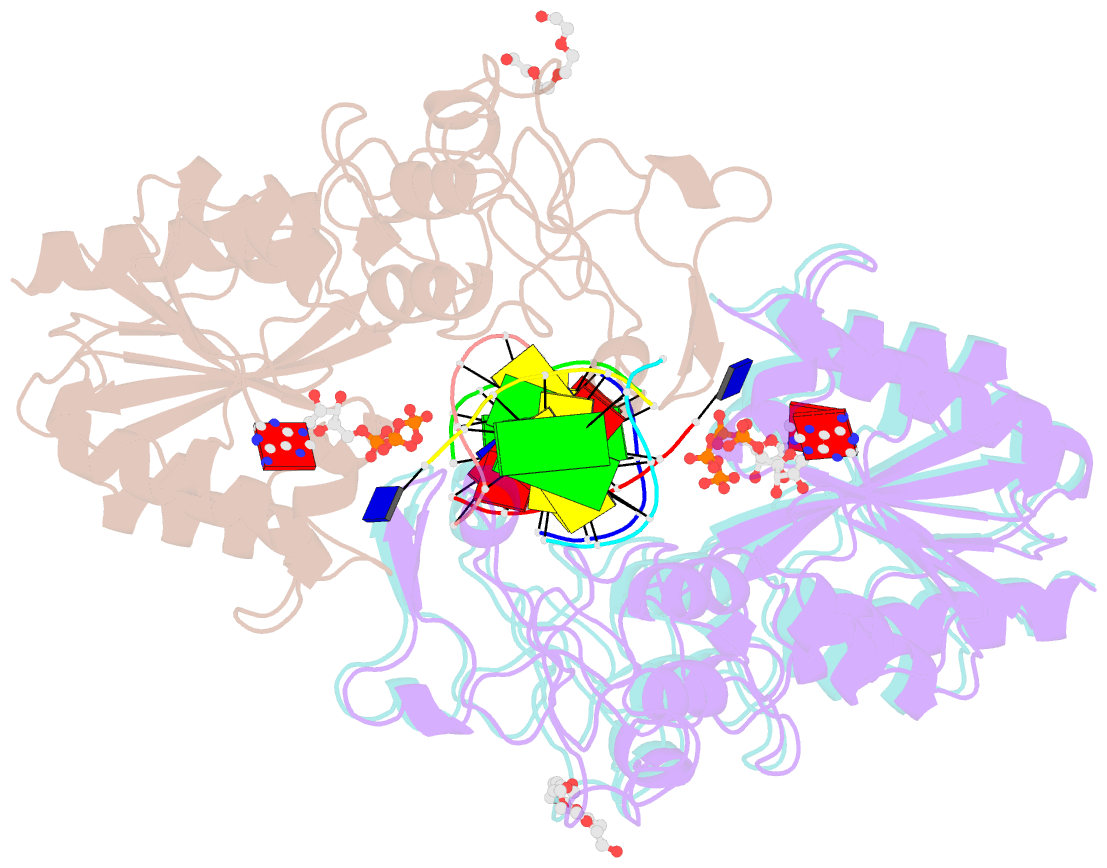
SNAP output for PDB entry 3ubt [SNAP web server]
Summary information and primary citation
- PDB-id
- 3ubt; SNAP-derived features in text and JSON formats; DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (2.502 Å)
- Summary
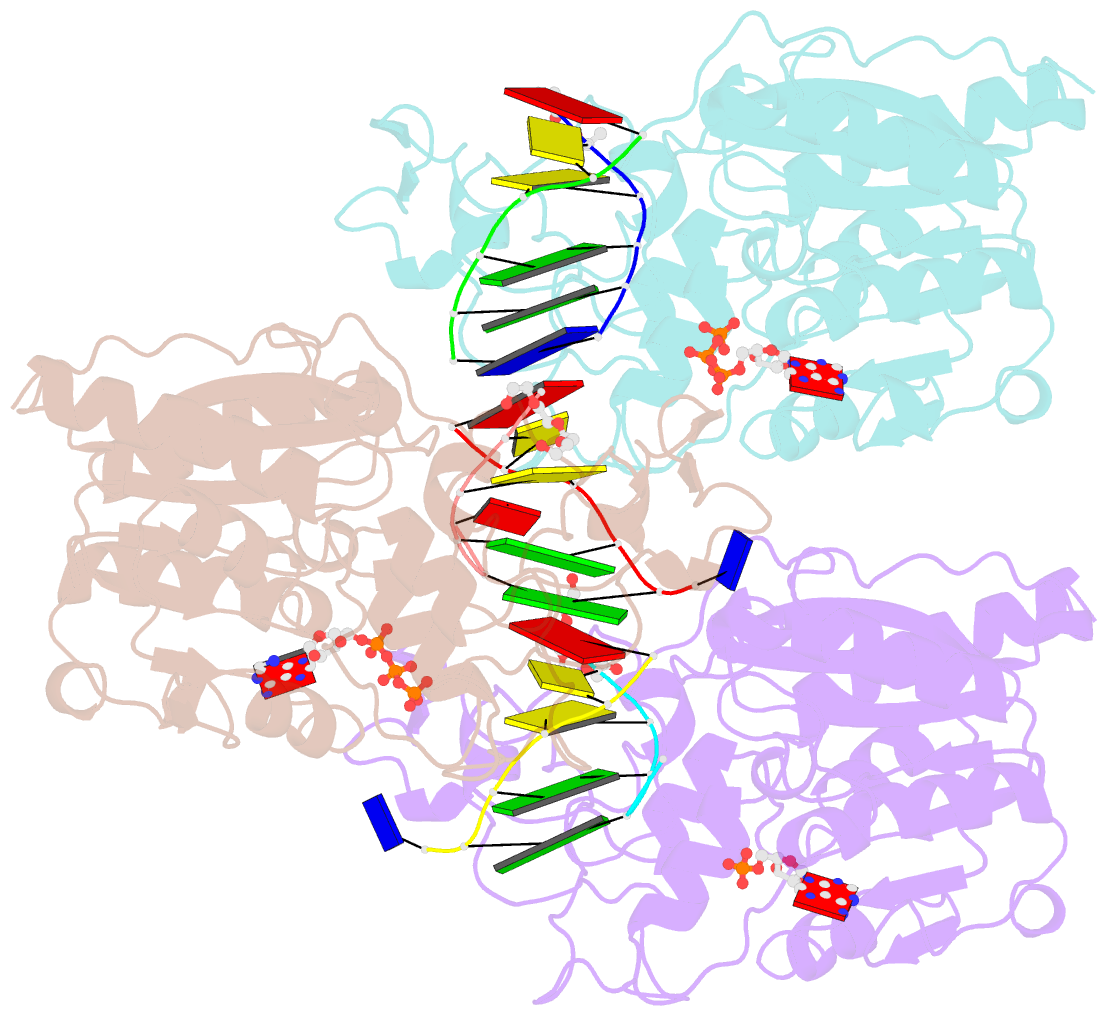
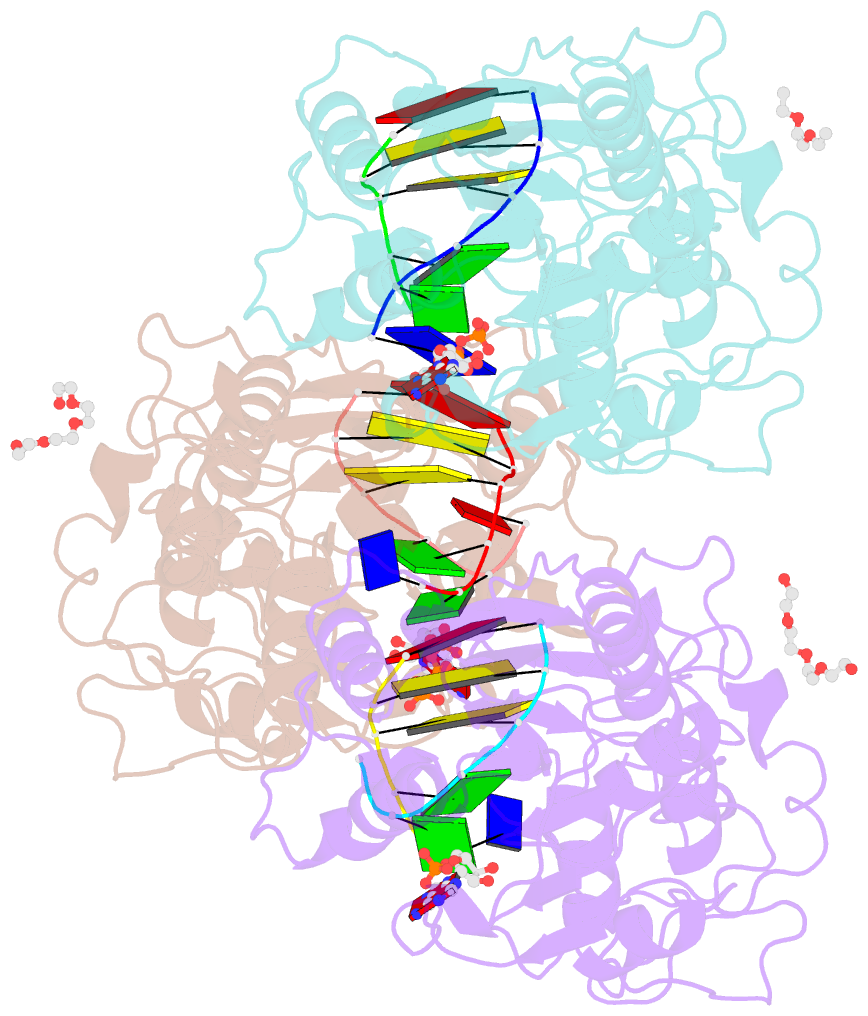
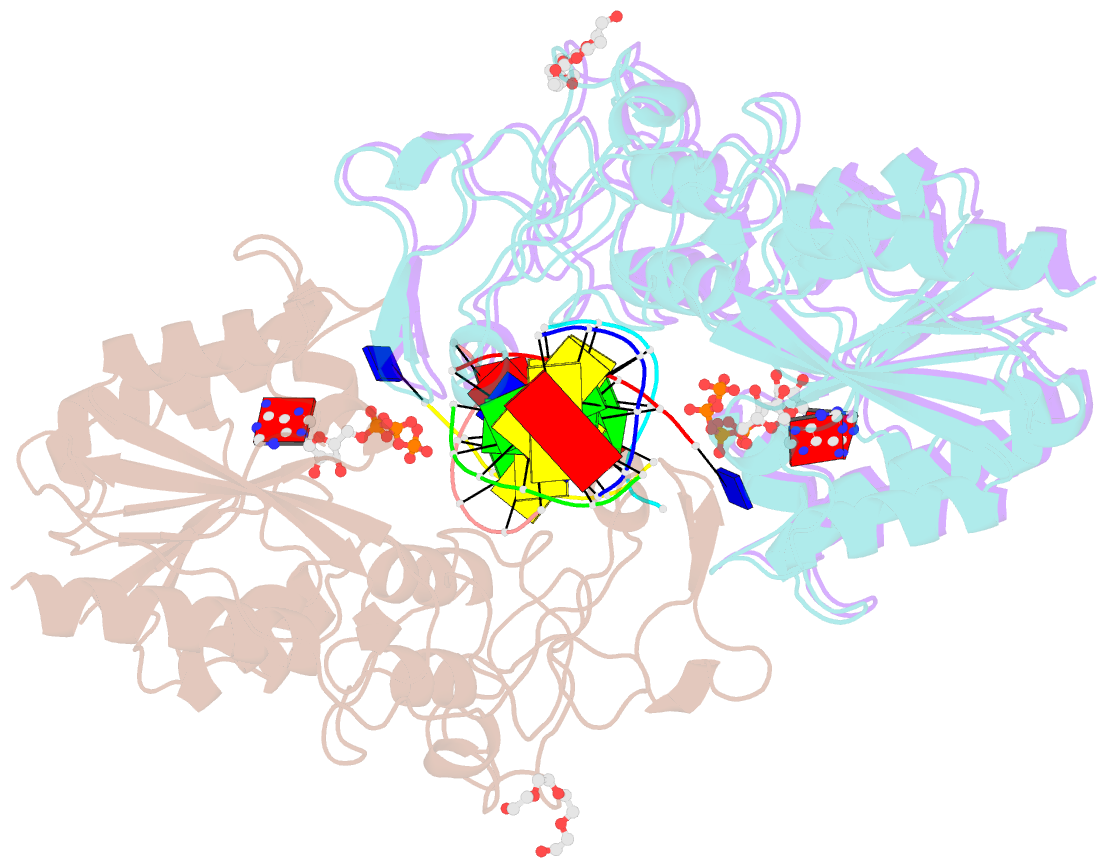
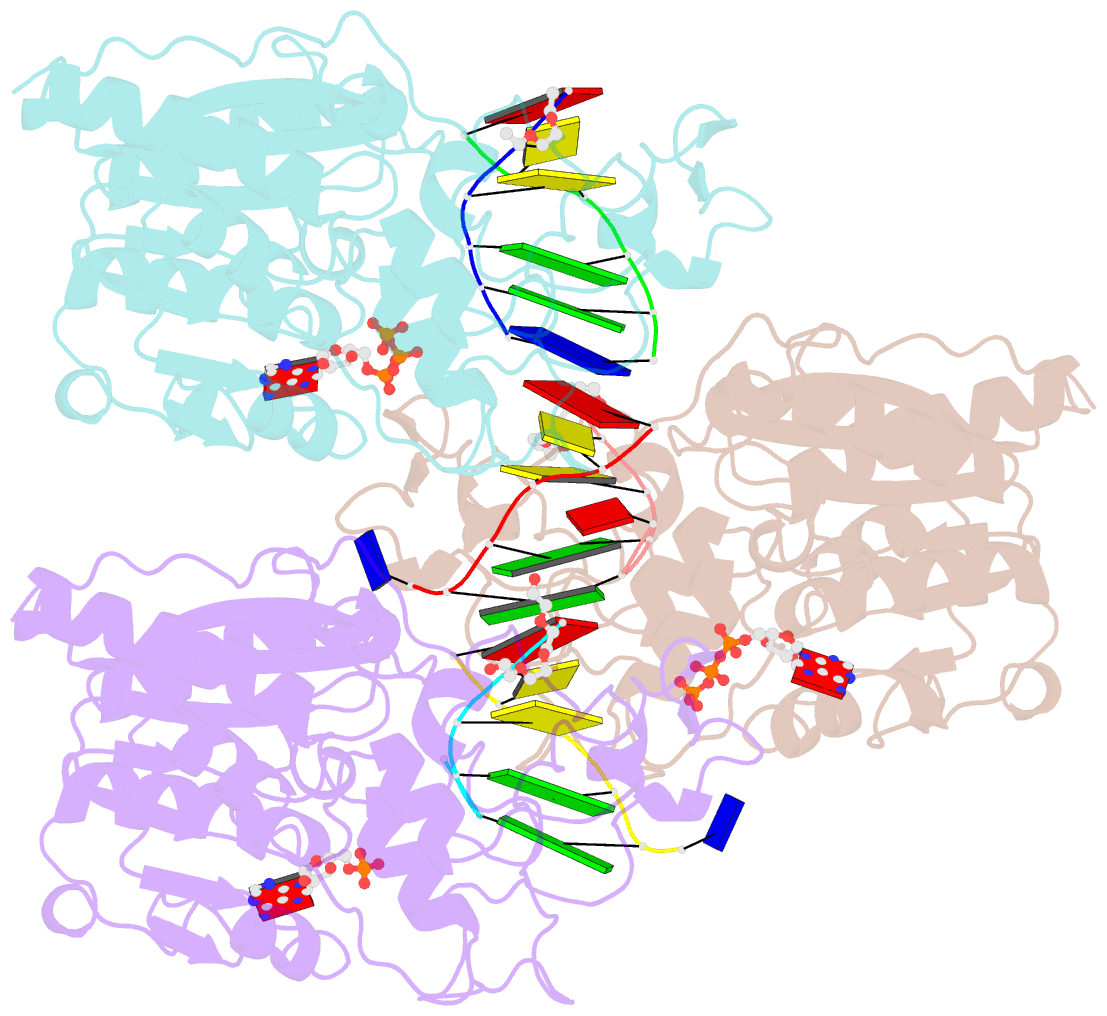
- Crystal structure of c71s mutant of DNA cytosine-5 methyltransferase m.haeiii bound to DNA
- Reference
- Didovyk A, Verdine GL (2012): "Structural origins of DNA target selection and nucleobase extrusion by a DNA Cytosine methyltransferase." J.Biol.Chem., 287, 40099-40105. doi: 10.1074/jbc.M112.413054.
- Abstract
- Background: How DNA 5-cytosine methyltransferases (DCMTases) select their substrate nucleobase for extrusion from DNA duplex is poorly understood.
Results: The crystal structure of a pre-extrusion M.HaeIII DCMTase-substrate DNA complex is reported here.