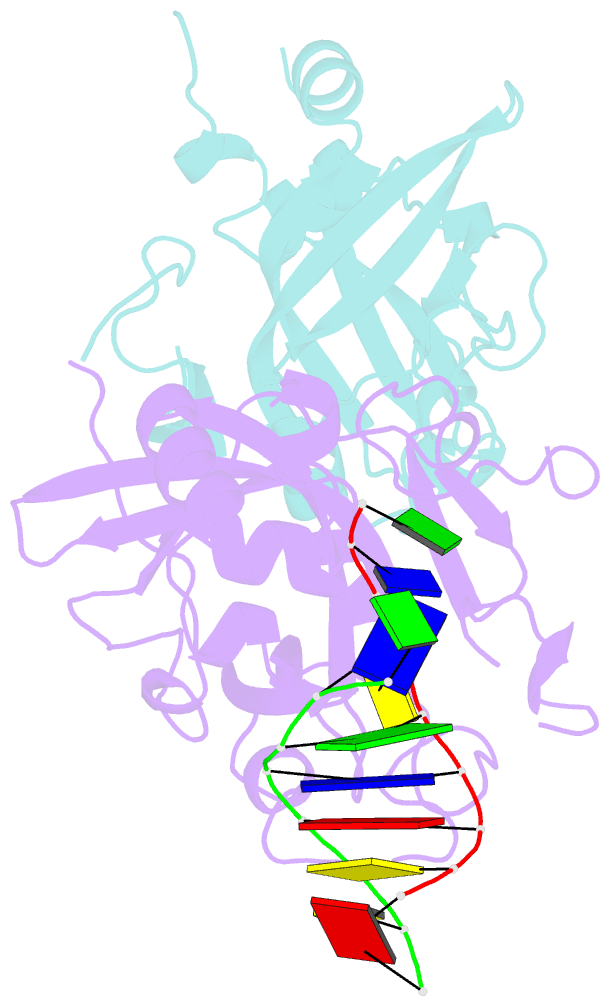
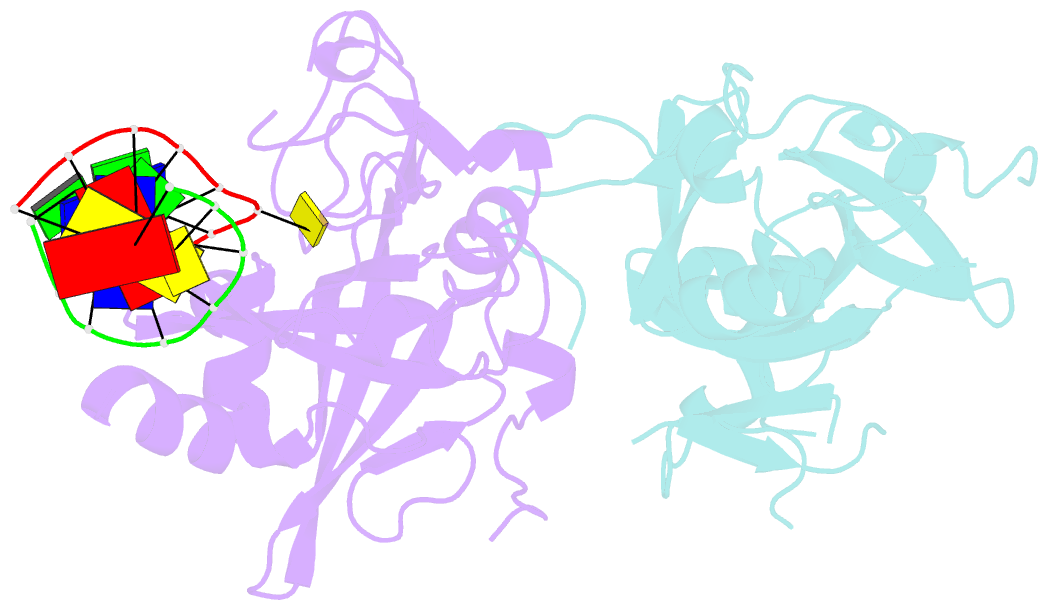
Summary information and primary citation
- PDB-id
- 3uby; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.0 Å)
- Summary
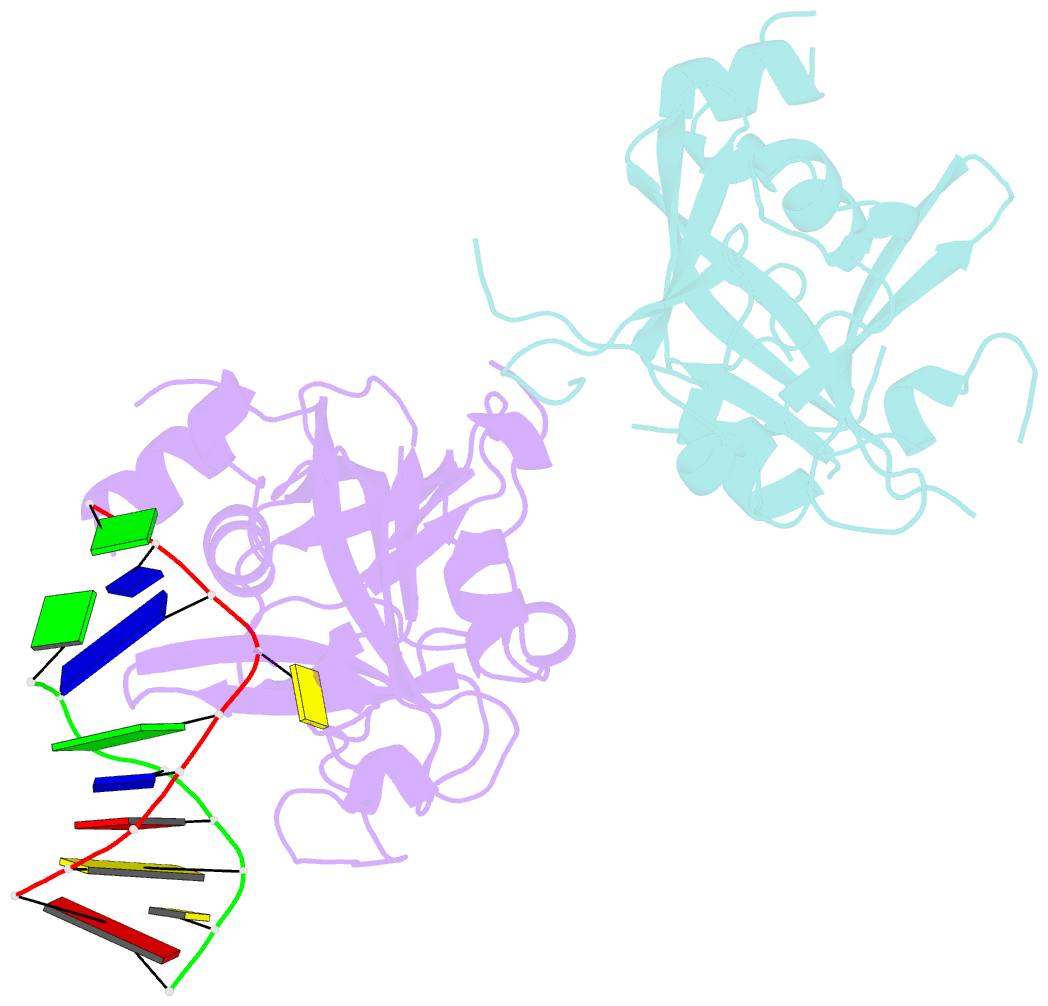
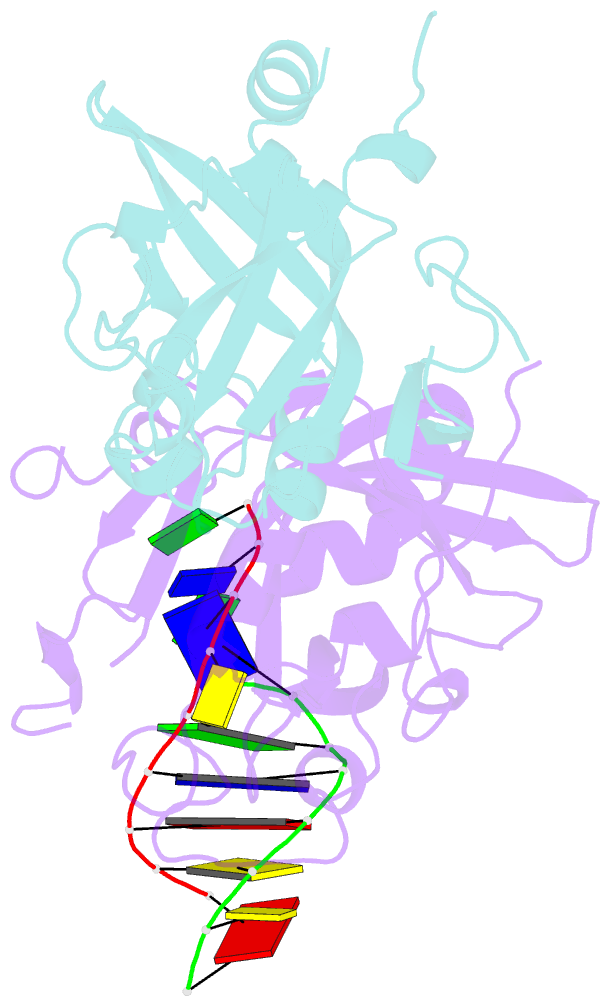
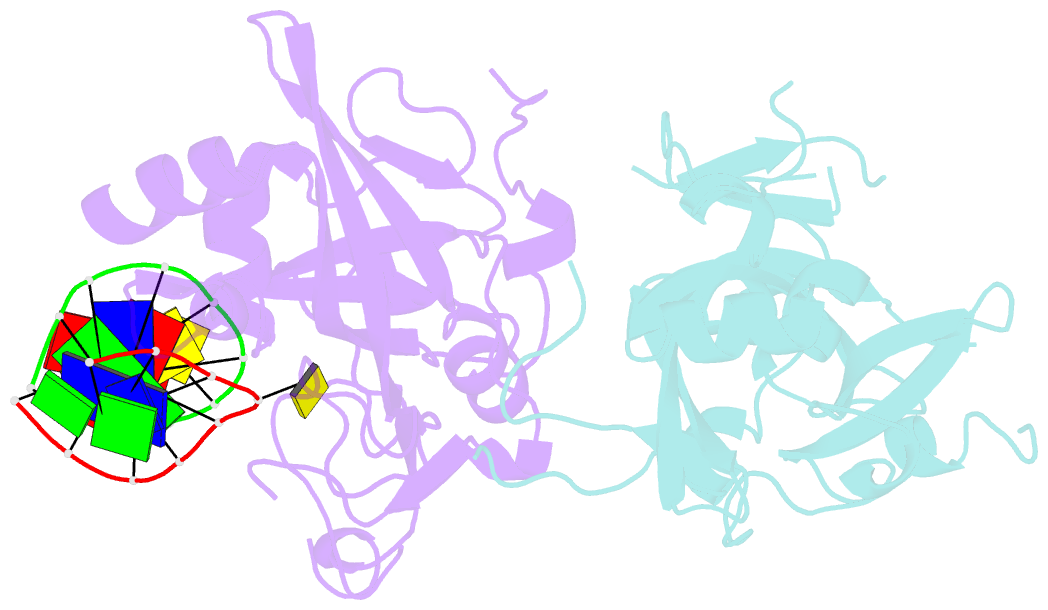
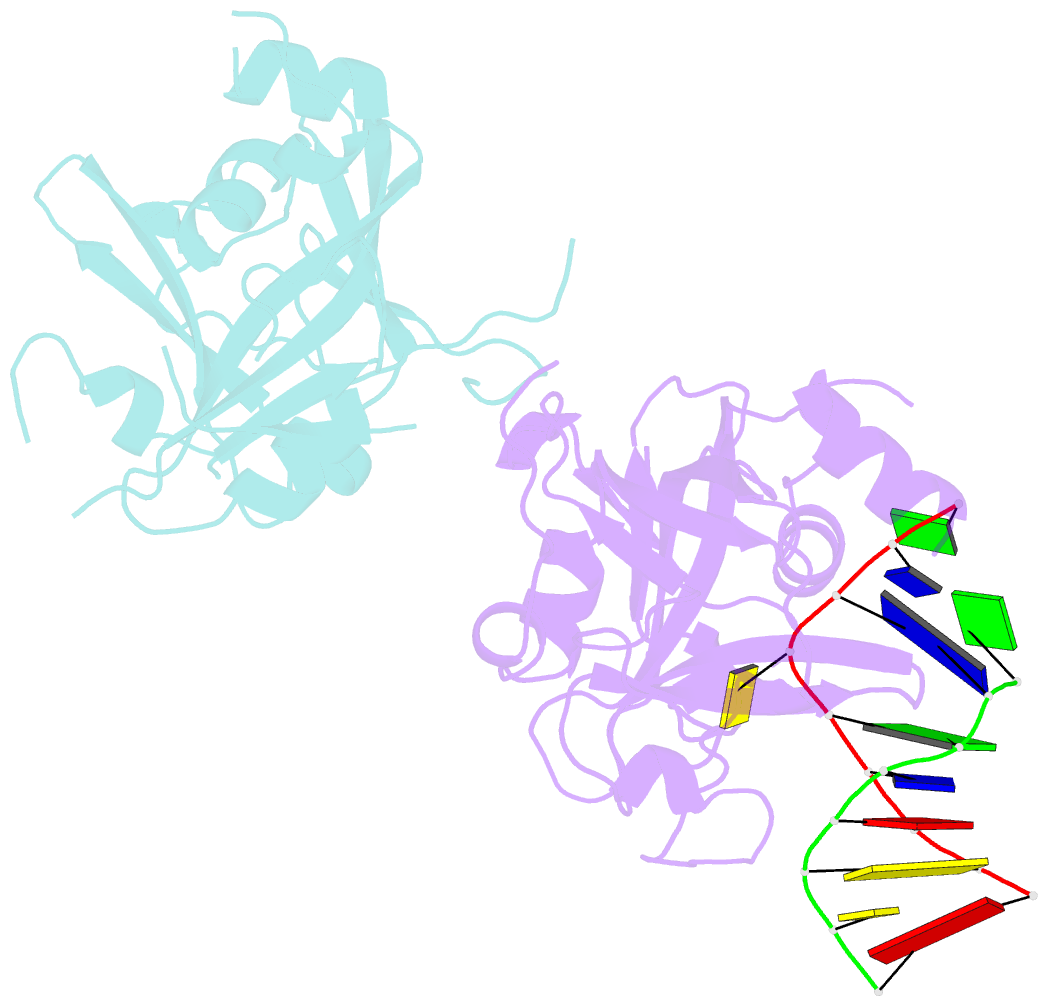
- Crystal structure of human alklyadenine DNA glycosylase in a lower and higher-affinity complex with DNA
- Reference
- Setser JW, Lingaraju GM, Davis CA, Samson LD, Drennan CL (2012): "Searching for DNA lesions: structural evidence for lower- and higher-affinity DNA binding conformations of human alkyladenine DNA glycosylase." Biochemistry, 51, 382-390. doi: 10.1021/bi201484k.
- Abstract
- To efficiently repair DNA, human alkyladenine DNA glycosylase (AAG) must search the million-fold excess of unmodified DNA bases to find a handful of DNA lesions. Such a search can be facilitated by the ability of glycosylases, like AAG, to interact with DNA using two affinities: a lower-affinity interaction in a searching process and a higher-affinity interaction for catalytic repair. Here, we present crystal structures of AAG trapped in two DNA-bound states. The lower-affinity depiction allows us to investigate, for the first time, the conformation of this protein in the absence of a tightly bound DNA adduct. We find that active site residues of AAG involved in binding lesion bases are in a disordered state. Furthermore, two loops that contribute significantly to the positive electrostatic surface of AAG are disordered. Additionally, a higher-affinity state of AAG captured here provides a fortuitous snapshot of how this enzyme interacts with a DNA adduct that resembles a one-base loop.