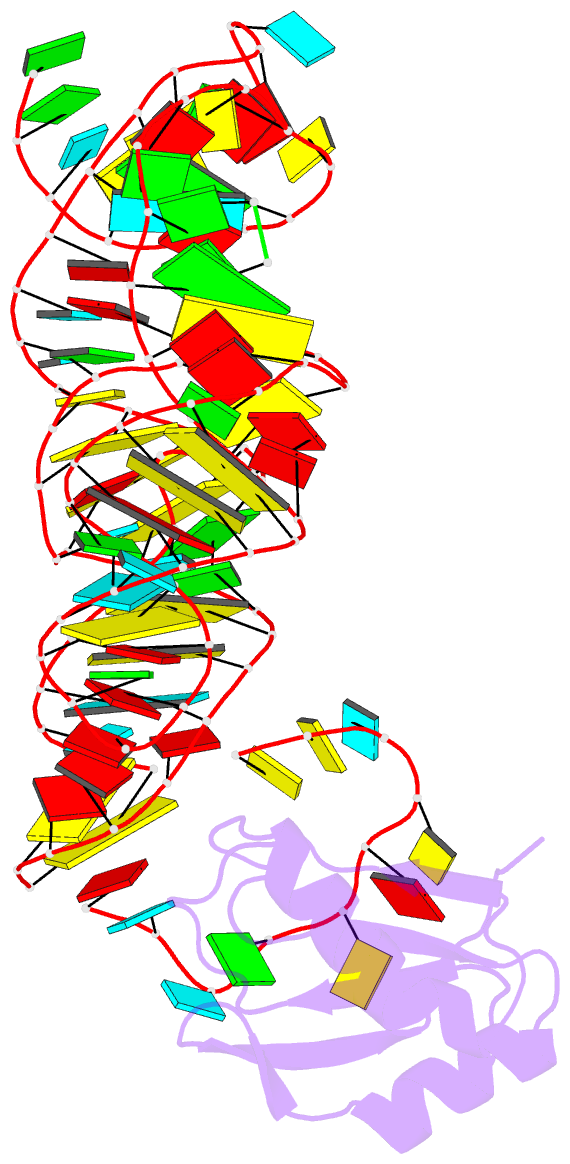
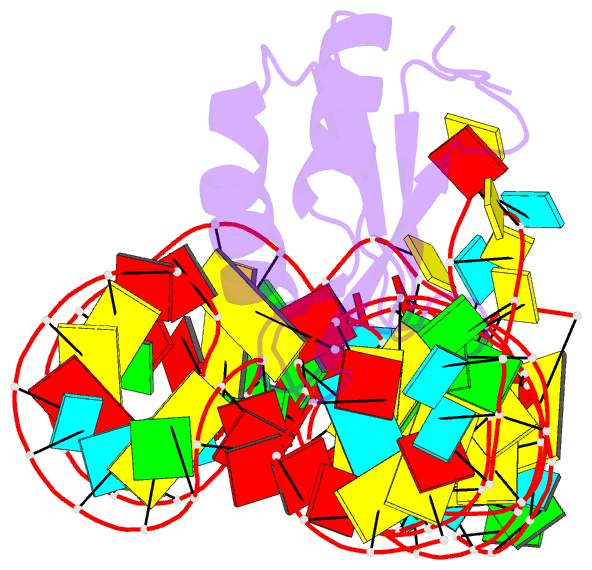
Summary information and primary citation
- PDB-id
- 3ud4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- signaling protein-RNA
- Method
- X-ray (2.7 Å)
- Summary
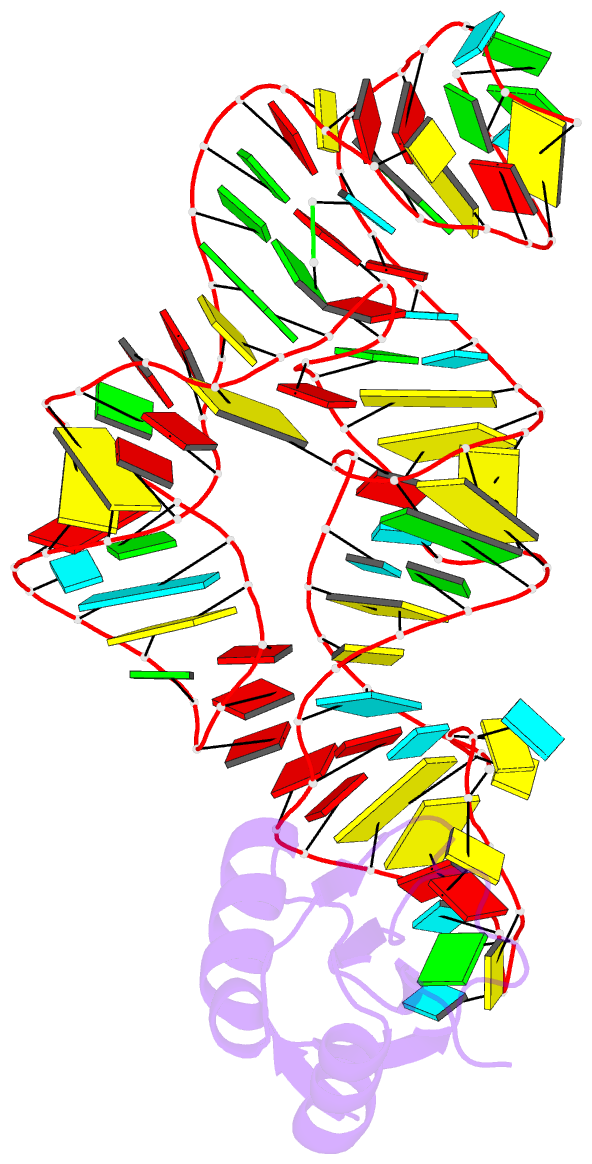
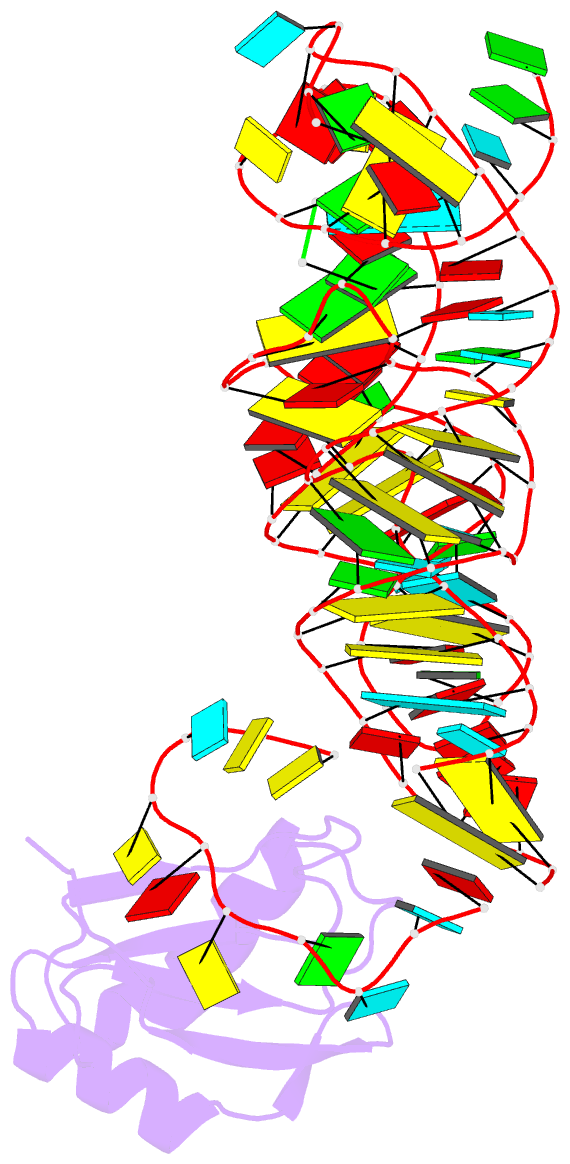
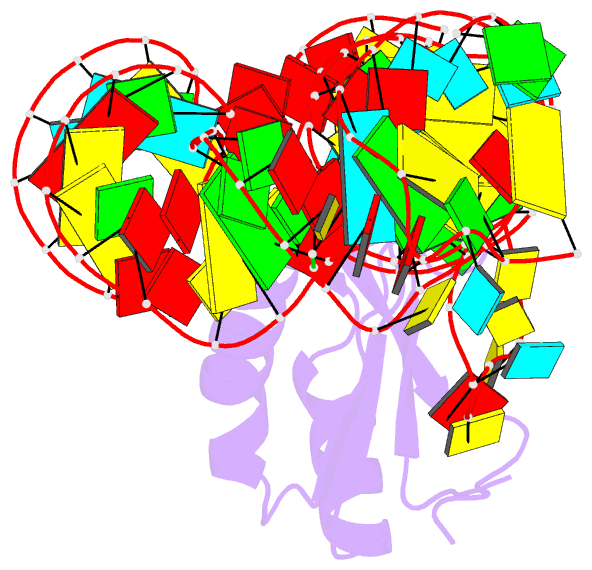
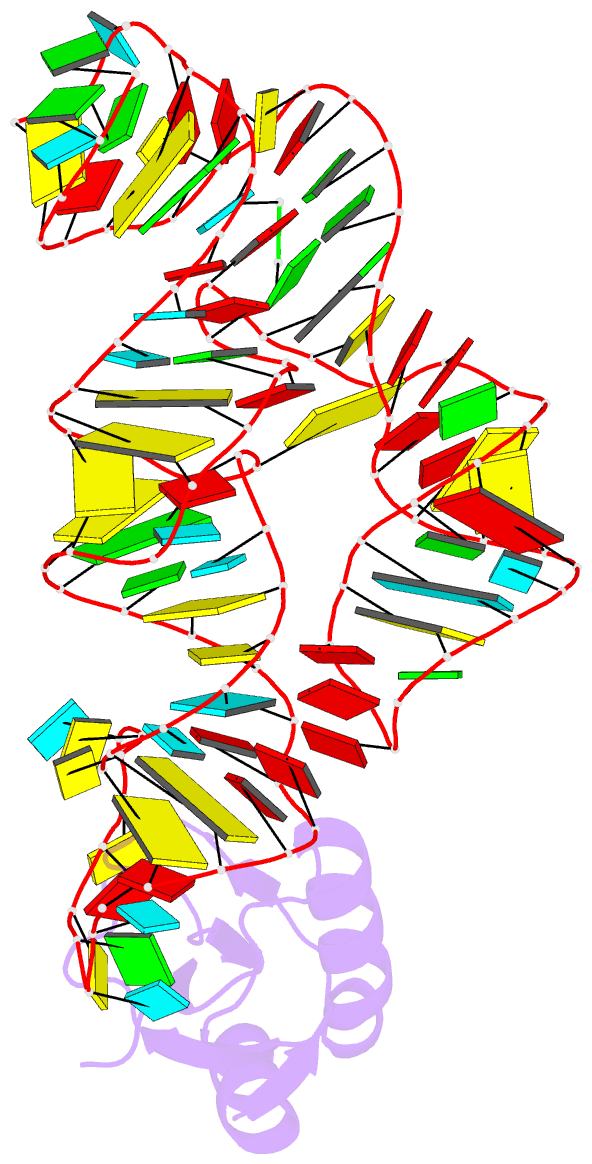
- The c92u mutant c-di-gmp-i riboswitch bound to gpa
- Reference
- Smith KD, Lipchock SV, Strobel SA (2012): "Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer." Biochemistry, 51, 425-432. doi: 10.1021/bi2016662.
- Abstract
- The cyclic dinucleotide c-di-GMP regulates lifestyle transitions in many bacteria, such as the change from a free motile state to a biofilm-forming community. Riboswitches that bind this second messenger are important downstream targets in this bacterial signaling pathway. The breakdown of c-di-GMP in the cell is accomplished enzymatically and results in the linear dinucleotide pGpG. The c-di-GMP-binding riboswitches must be able to discriminate between their cognate cyclic ligand and linear dinucleotides in order to be selective biological switches. It has been reported that the c-di-GMP-I riboswitch binds c-di-GMP 5 orders of magnitude better than the linear pGpG, but the cause of this large energetic difference in binding is unknown. Here we report binding data and crystal structures of several linear c-di-GMP analogues in complex with the c-di-GMP-I riboswitch. These data reveal the parameters for phosphate recognition and the structural basis of linear dinucleotide binding to the riboswitch. Additionally, the pH dependence of binding shows that exclusion of pGpG is not due to the additional negative charge on the ligand. These data reveal principles that, along with published work, will contribute to the design of c-di-GMP analogues with properties desirable for use as chemical tools and potential therapeutics.