Summary information and primary citation
- PDB-id
- 3ugp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (2.697 Å)
- Summary
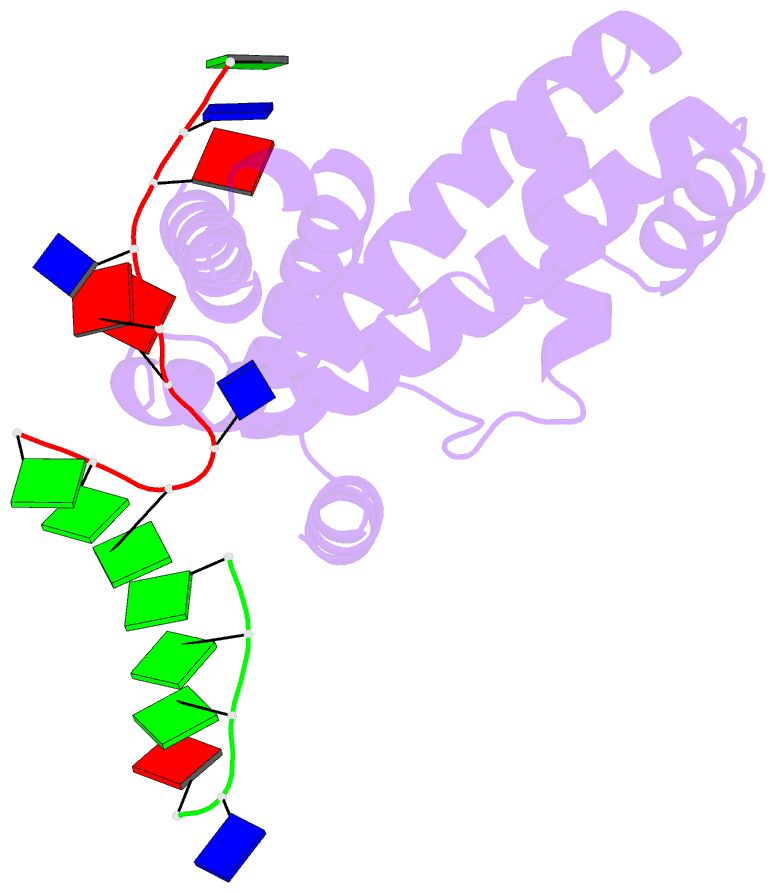
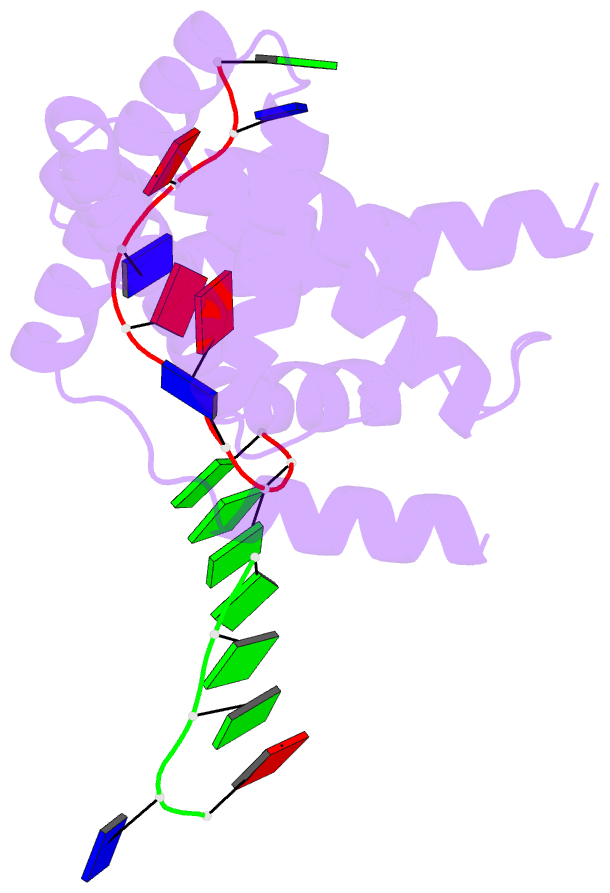
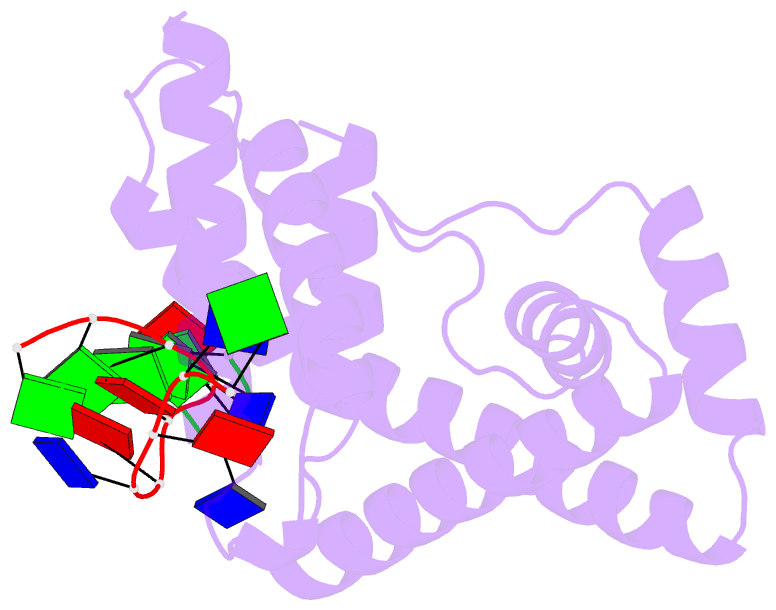
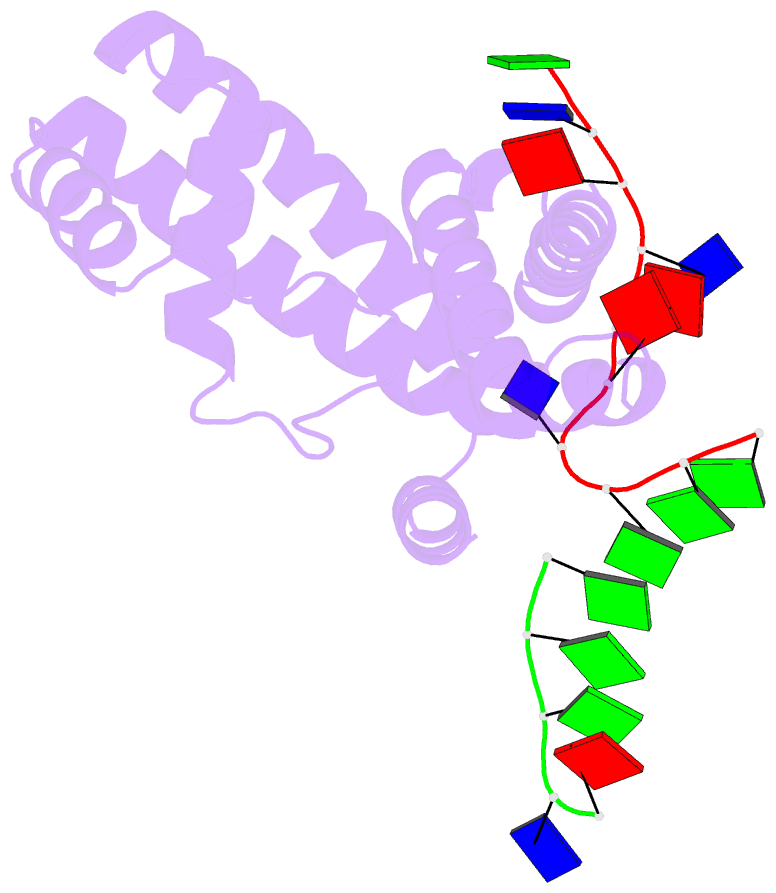
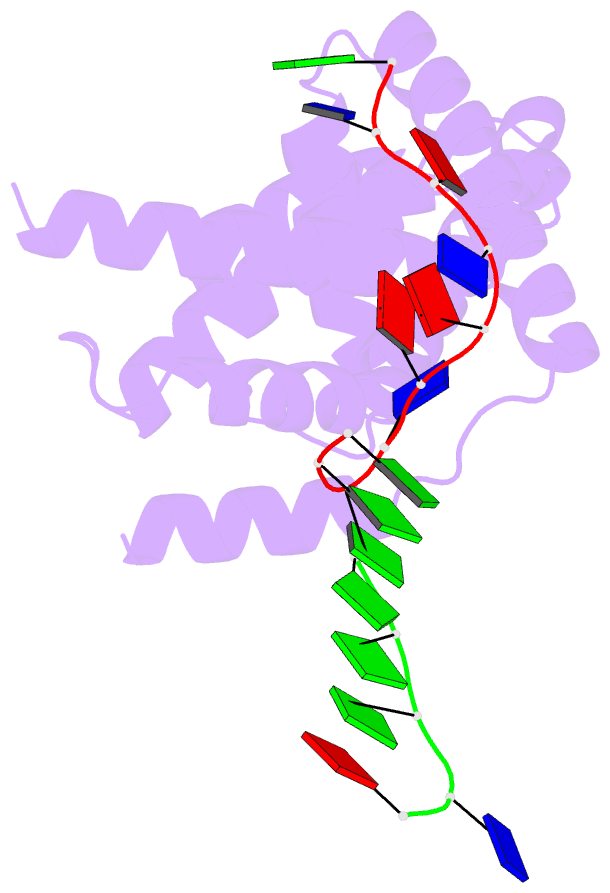
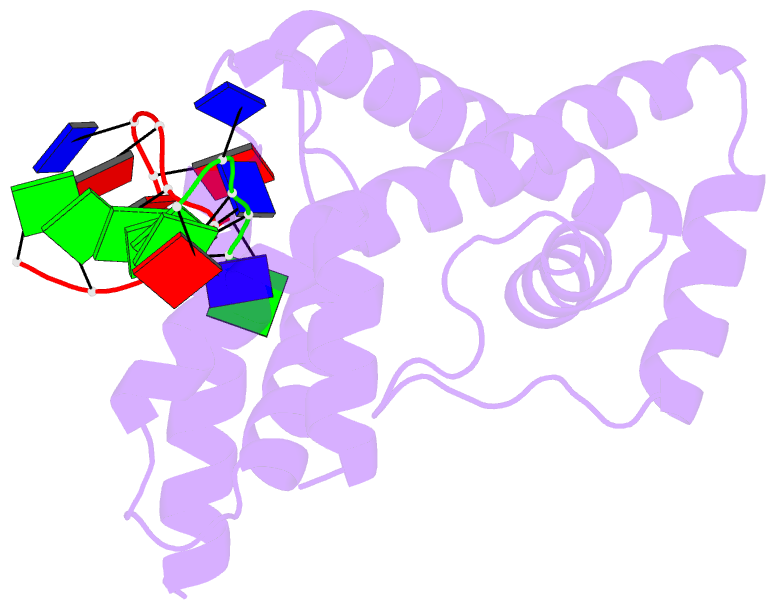
- Crystal structure of RNA-polymerase sigma subunit domain 2 complexed with -10 promoter element ssDNA oligo (tataat)
- Reference
- Feklistov A, Darst SA (2011): "Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit." Cell(Cambridge,Mass.), 147, 1257-1269. doi: 10.1016/j.cell.2011.10.041.
- Abstract
- The key step in bacterial promoter opening is recognition of the -10 promoter element (T(-12)A(-11)T(-10)A(-9)A(-8)T(-7) consensus sequence) by the RNA polymerase σ subunit. We determined crystal structures of σ domain 2 bound to single-stranded DNA bearing-10 element sequences. Extensive interactions occur between the protein and the DNA backbone of every -10 element nucleotide. Base-specific interactions occur primarily with A(-11) and T(-7), which are flipped out of the single-stranded DNA base stack and buried deep in protein pockets. The structures, along with biochemical data, support a model where the recognition of the -10 element sequence drives initial promoter opening as the bases of the nontemplate strand are extruded from the DNA double-helix and captured by σ. These results provide a detailed structural basis for the critical roles of A(-11) and T(-7) in promoter melting and reveal important insights into the initiation of transcription bubble formation.