Summary information and primary citation
- PDB-id
- 3upu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.299 Å)
- Summary
- Crystal structure of the t4 phage sf1b helicase dda
- Reference
- He X, Byrd AK, Yun MK, Pemble CW, Harrison D, Yeruva L, Dahl C, Kreuzer KN, Raney KD, White SW (2012): "The T4 Phage SF1B Helicase Dda Is Structurally Optimized to Perform DNA Strand Separation." Structure, 20, 1189-1200. doi: 10.1016/j.str.2012.04.013.
- Abstract
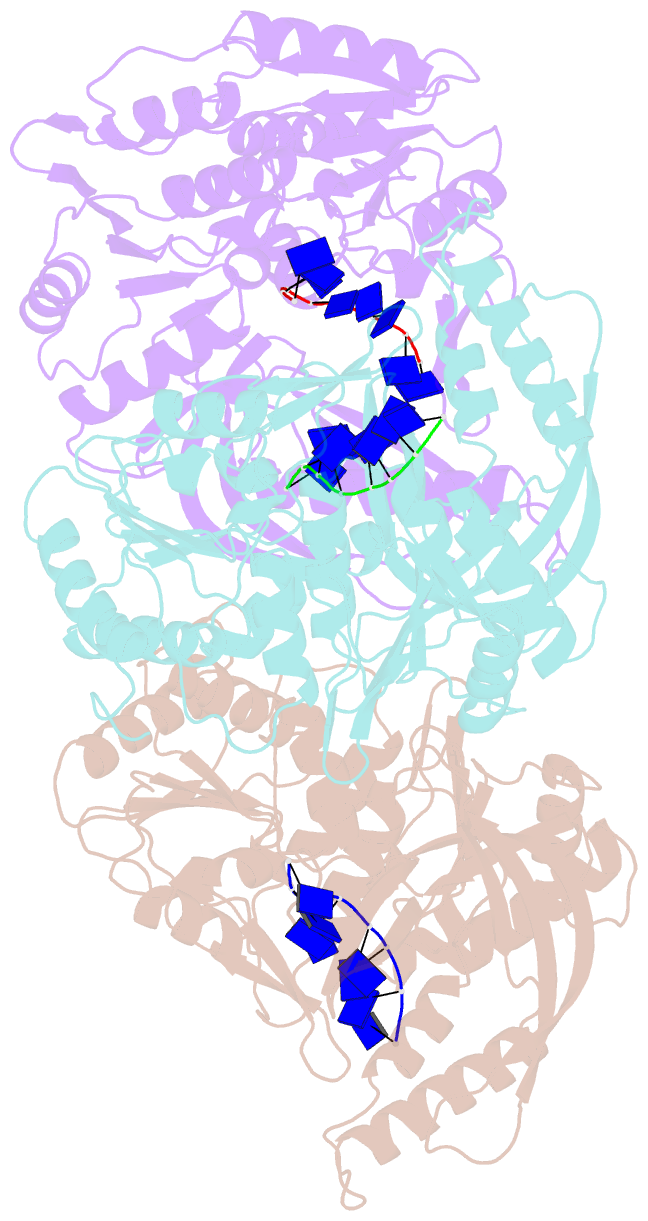
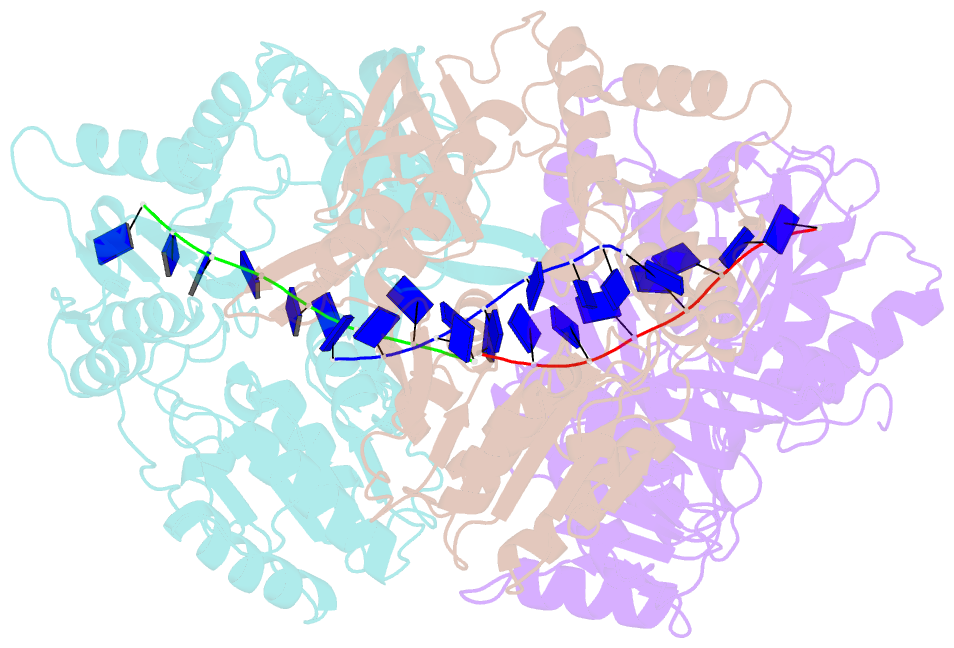
- Helicases move on DNA via an ATP binding and hydrolysis mechanism coordinated by well-characterized helicase motifs. However, the translocation along single-stranded DNA (ssDNA) and the strand separation of double-stranded (dsDNA) may be loosely or tightly coupled. Dda is a phage T4 SF1B helicase with sequence homology to the Pif1 family of helicases that tightly couples translocation to strand separation. The crystal structure of the Dda-ssDNA binary complex reveals a domain referred to as the "pin" that was previously thought to remain static during strand separation. The pin contains a conserved phenylalanine that mediates a transient base-stacking interaction that is absolutely required for separation of dsDNA. The pin is secured at its tip by protein-protein interactions through an extended SH3 domain thereby creating a rigid strut. The conserved interface between the pin and the SH3 domain provides the mechanism for tight coupling of translocation to strand separation.