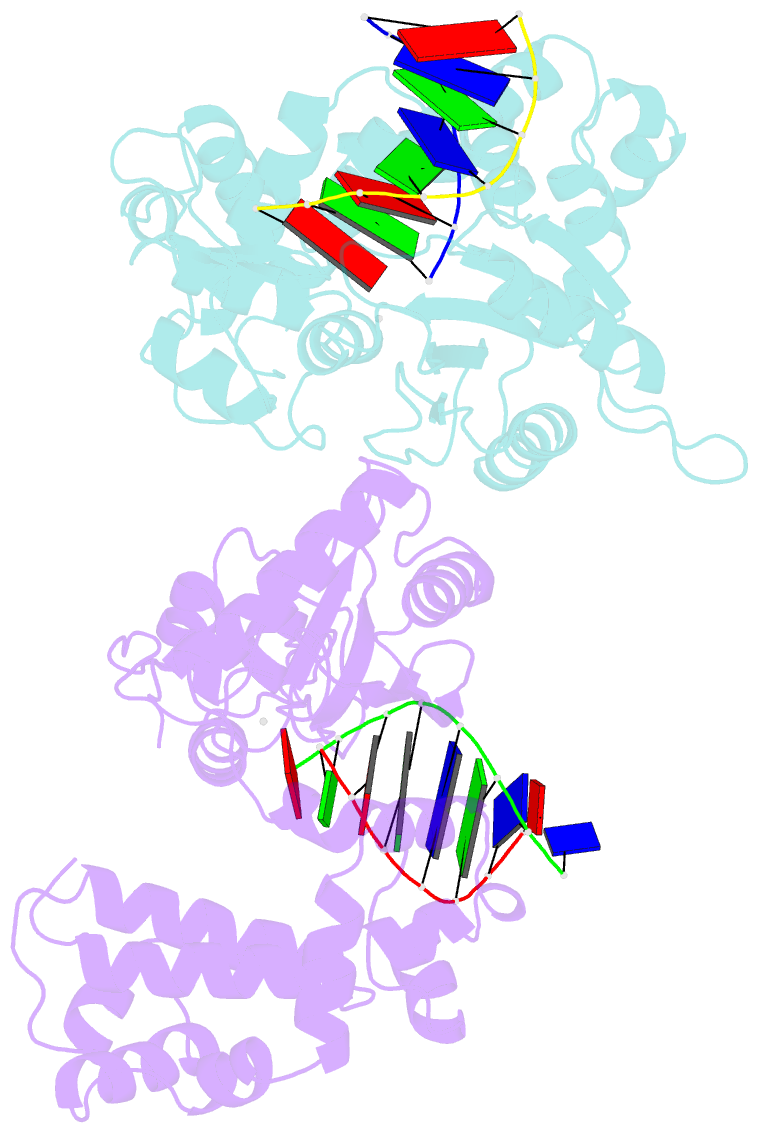
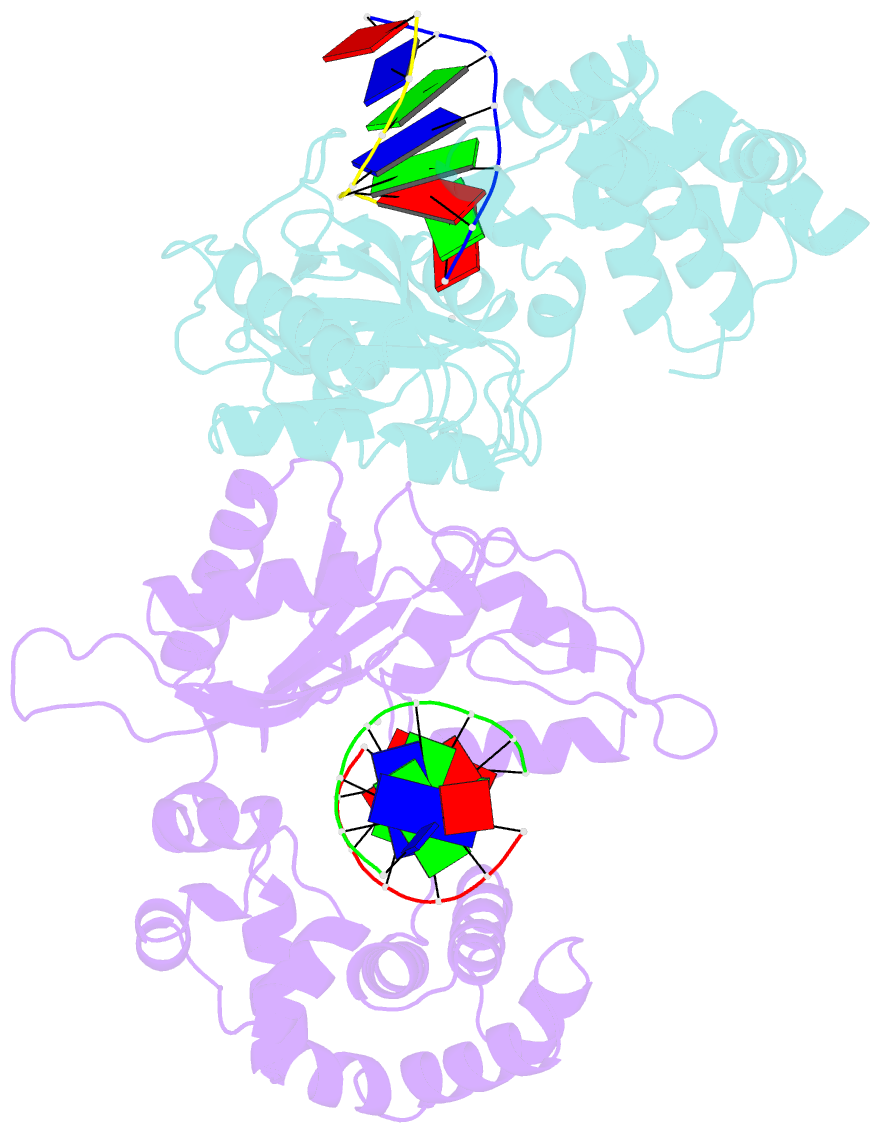
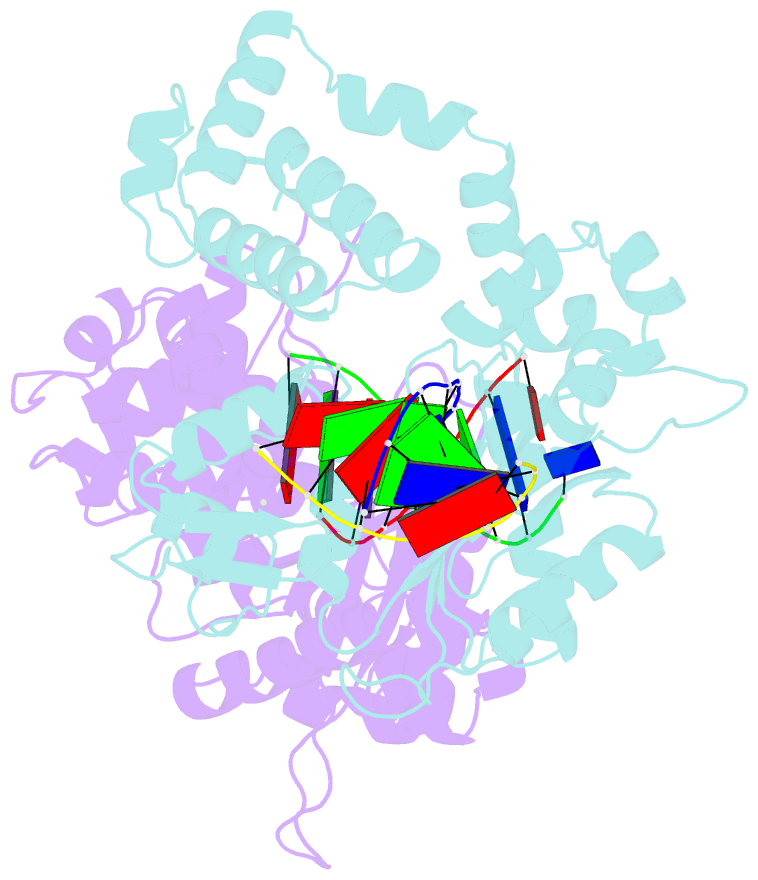
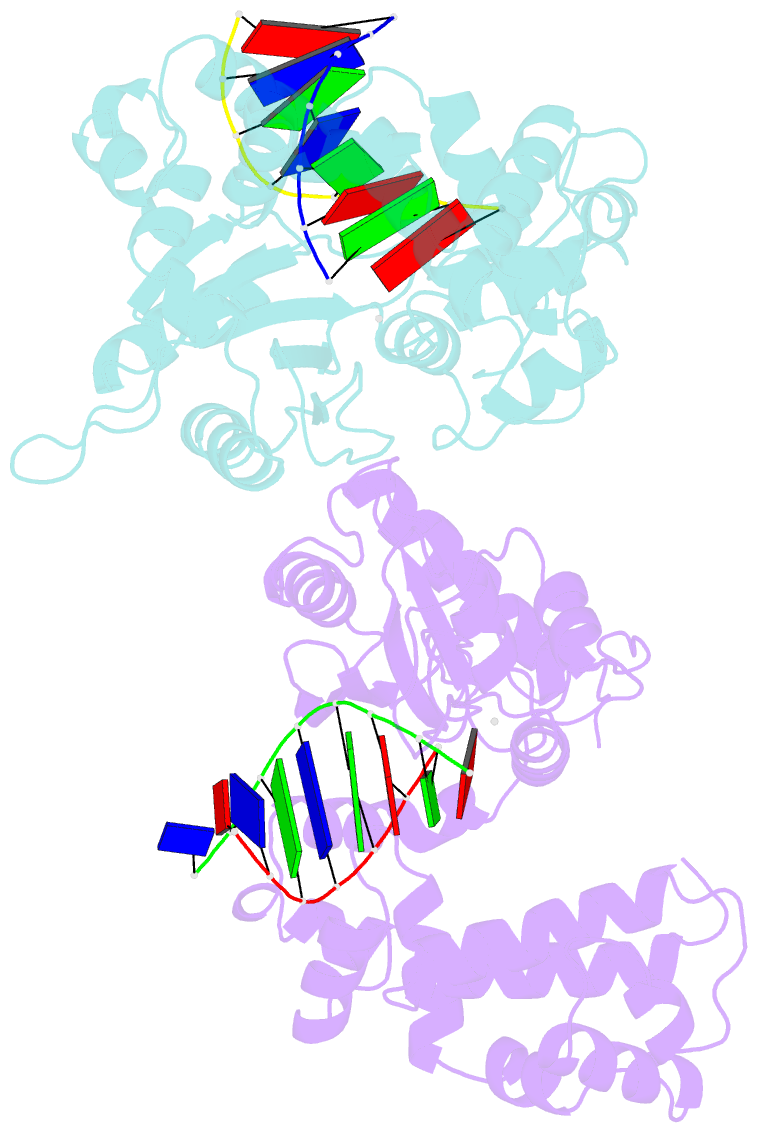
Summary information and primary citation
- PDB-id
- 3uxp; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase, lyase-DNA
- Method
- X-ray (2.723 Å)
- Summary
- Co-crystal structure of rat DNA polymerase beta mutator i260q: enzyme-DNA-ddttp
- Reference
- Gridley CL, Rangarajan S, Firbank S, Dalal S, Sweasy JB, Jaeger J (2013): "Structural Changes in the Hydrophobic Hinge Region Adversely Affect the Activity and Fidelity of the I260Q Mutator DNA Polymerase beta." Biochemistry, 52, 4422-4432. doi: 10.1021/bi301368f.
- Abstract
- The I260Q variant of DNA polymerase β is an efficient mutator polymerase with fairly indiscriminate misincorporation activities opposite all template bases. Previous modeling studies have suggested that I260Q harbors structural variations in its hinge region. Here, we present the crystal structures of wild type and I260Q rat polymerase β in the presence and absence of substrates. Both the I260Q apoenzyme structure and the closed ternary complex with double-stranded DNA and ddTTP show ordered water molecules in the hydrophobic hinge near Gln260, whereas this is not the case in the wild type polymerase. Compared to wild type polymerase β ternary complexes, there are subtle movements around residues 260, 272, 295, and 296 in the mutant. The rearrangements in this region, coupled with side chain movements in the immediate neighborhood of the dNTP-binding pocket, namely, residues 258 and 272, provide an explanation for the altered activity and fidelity profiles observed in the I260Q mutator polymerase.