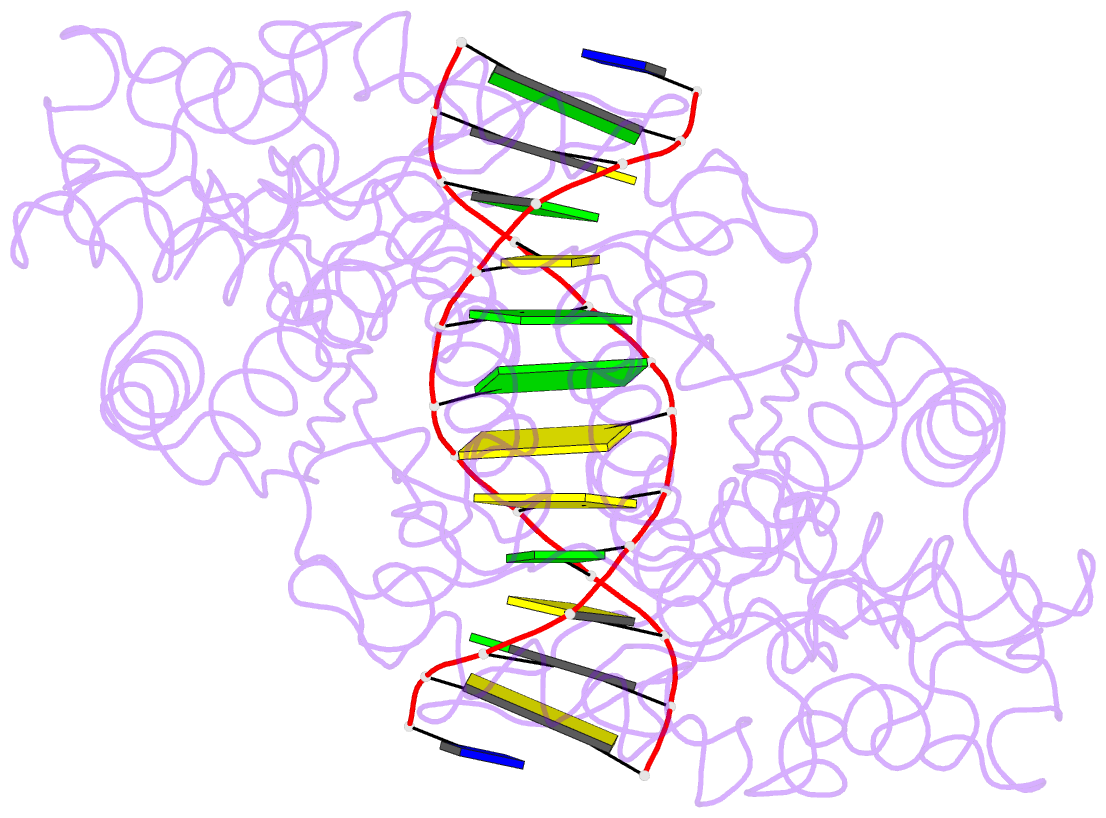
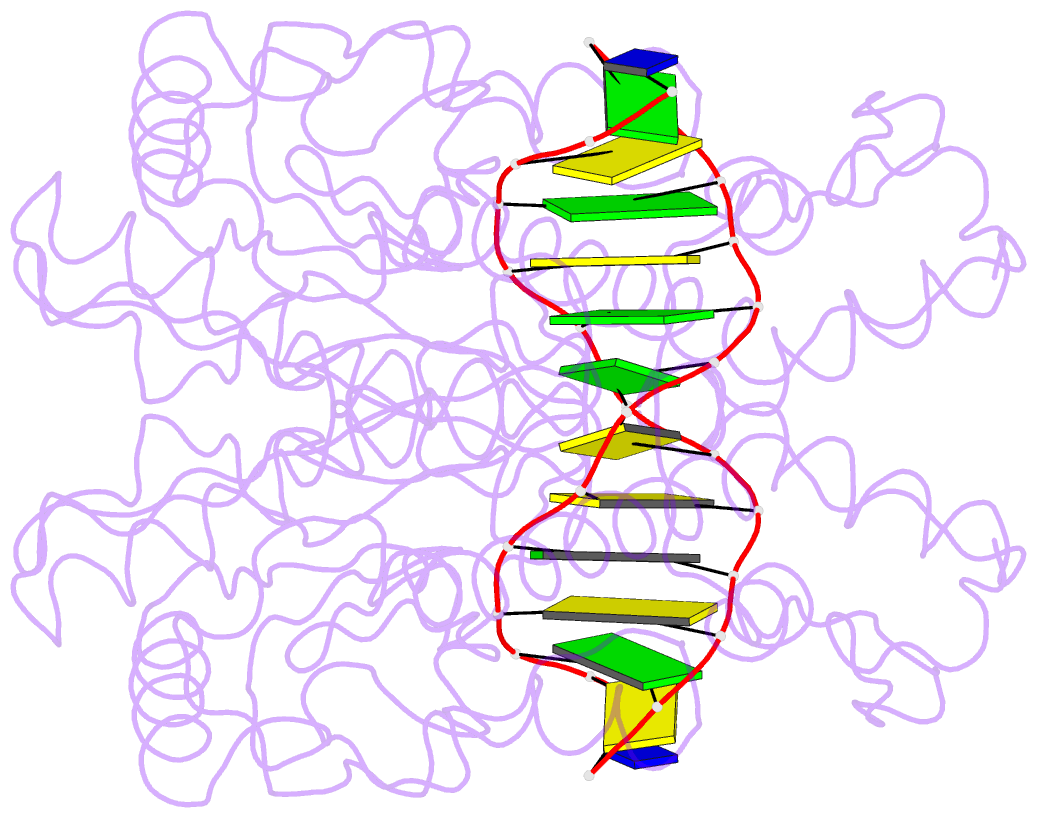
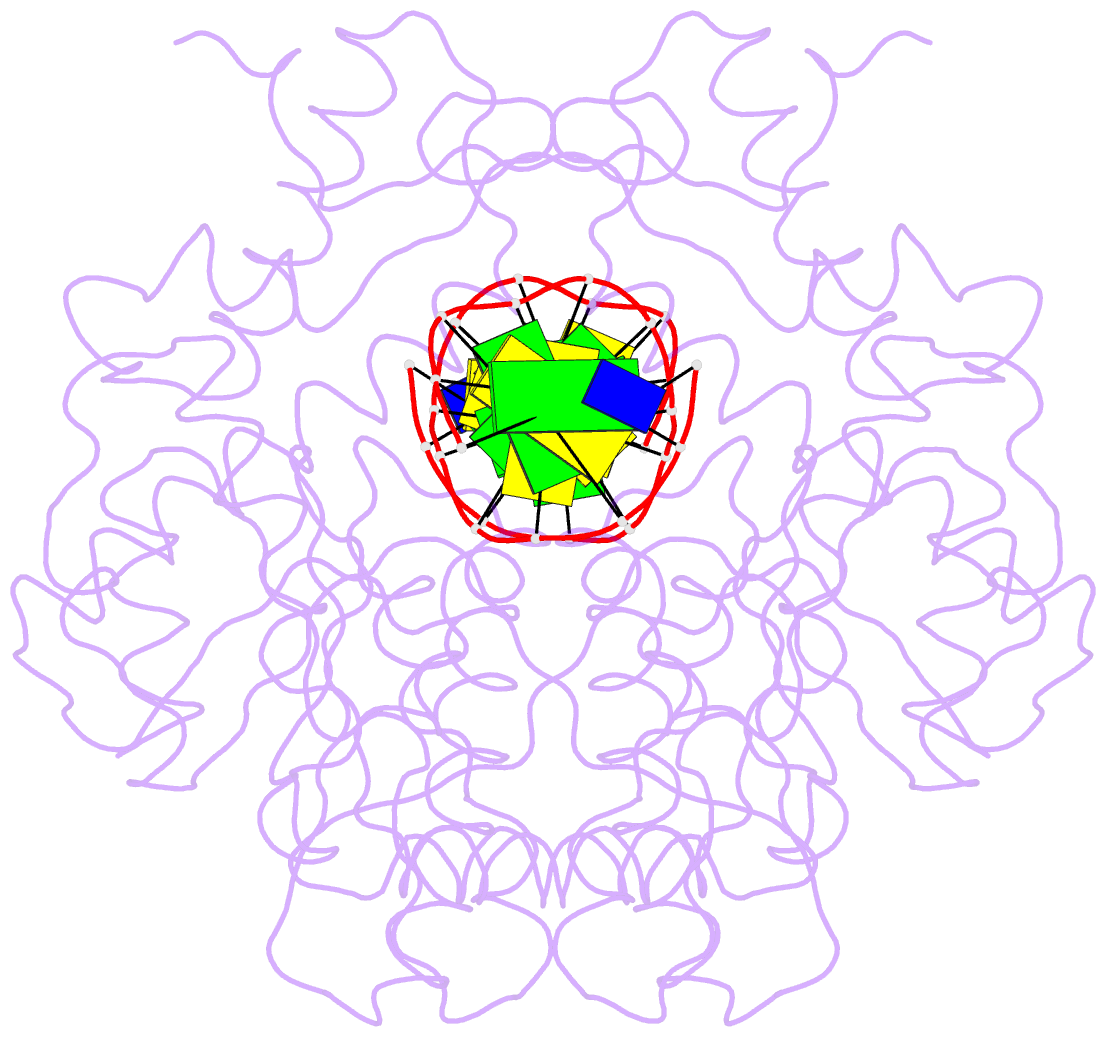
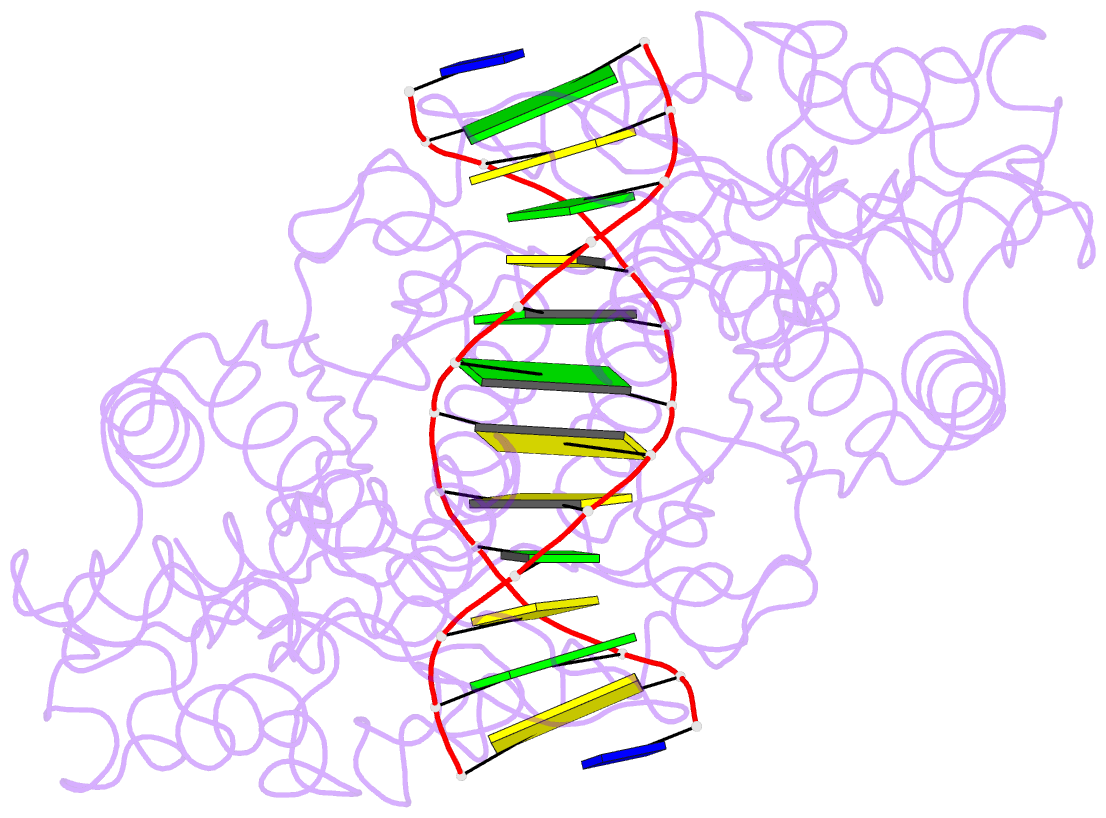
Summary information and primary citation
- PDB-id
- 3v1z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.2 Å)
- Summary
- Crystal structure of type iif restriction endonuclease bse634i with cognate DNA
- Reference
- Manakova E, Grazulis S, Zaremba M, Tamulaitiene G, Golovenko D, Siksnys V (2012): "Structural mechanisms of the degenerate sequence recognition by Bse634I restriction endonuclease." Nucleic Acids Res., 40, 6741-6751. doi: 10.1093/nar/gks300.
- Abstract
- Restriction endonuclease Bse634I recognizes and cleaves the degenerate DNA sequence 5'-R/CCGGY-3' (R stands for A or G; Y for T or C, '/' indicates a cleavage position). Here, we report the crystal structures of the Bse634I R226A mutant complexed with cognate oligoduplexes containing ACCGGT and GCCGGC sites, respectively. In the crystal, all potential H-bond donor and acceptor atoms on the base edges of the conserved CCGG core are engaged in the interactions with Bse634I amino acid residues located on the α6 helix. In contrast, direct contacts between the protein and outer base pairs are limited to van der Waals contact between the purine nucleobase and Pro203 residue in the major groove and a single H-bond between the O2 atom of the outer pyrimidine and the side chain of the Asn73 residue in the minor groove. Structural data coupled with biochemical experiments suggest that both van der Waals interactions and indirect readout contribute to the discrimination of the degenerate base pair by Bse634I. Structure comparison between related enzymes Bse634I (R/CCGGY), NgoMIV (G/CCGGC) and SgrAI (CR/CCGGYG) reveals how different specificities are achieved within a conserved structural core.