Summary information and primary citation
- PDB-id
- 3va0; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.201 Å)
- Summary
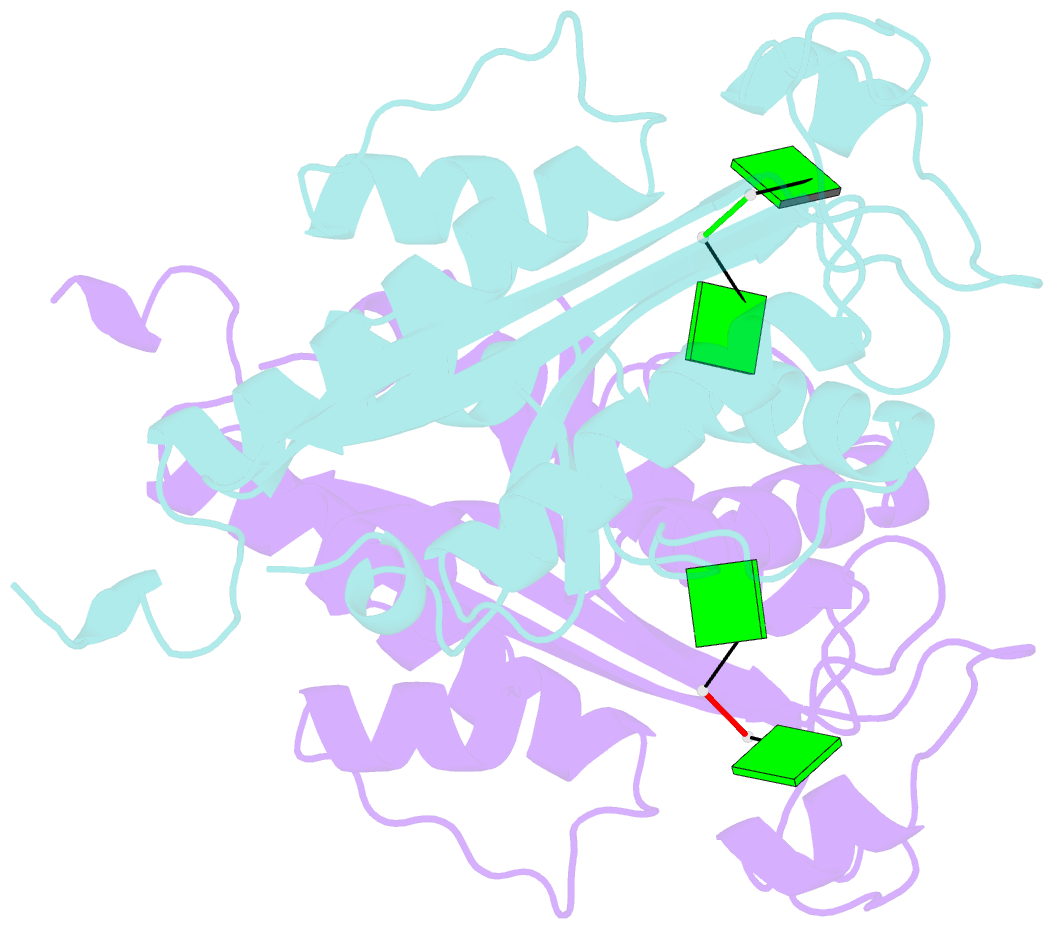
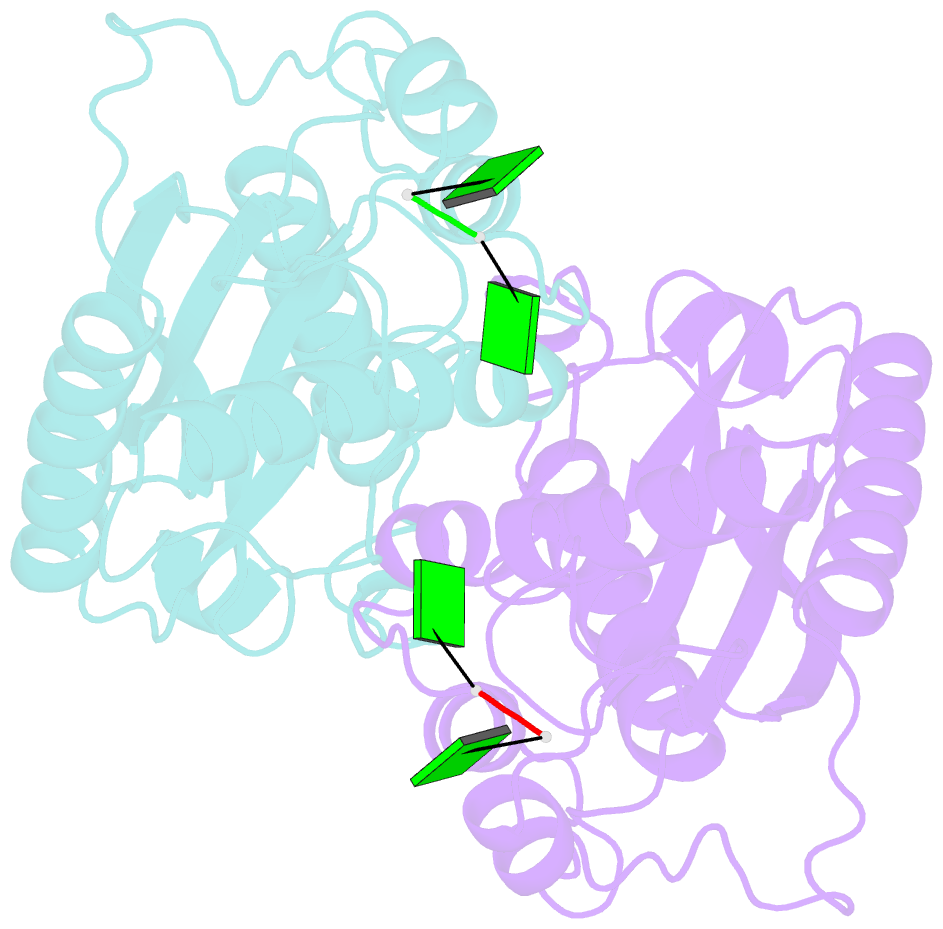
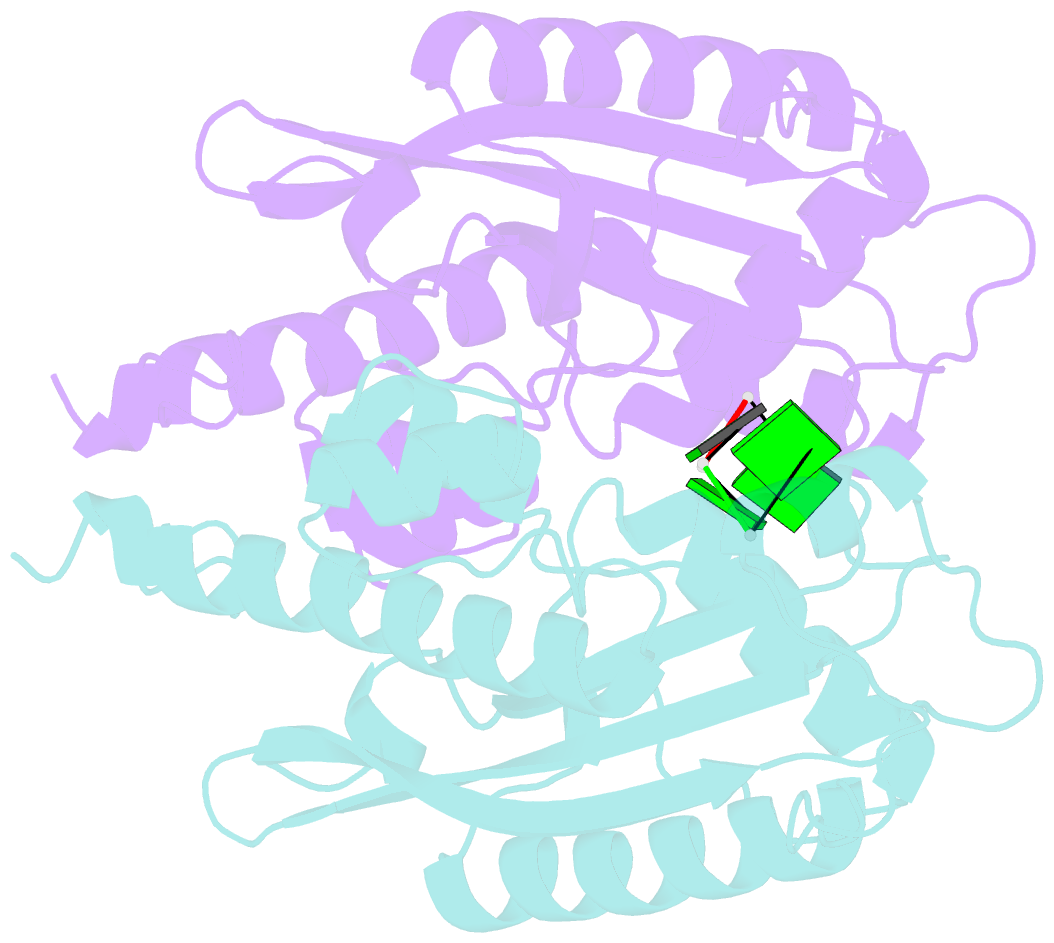
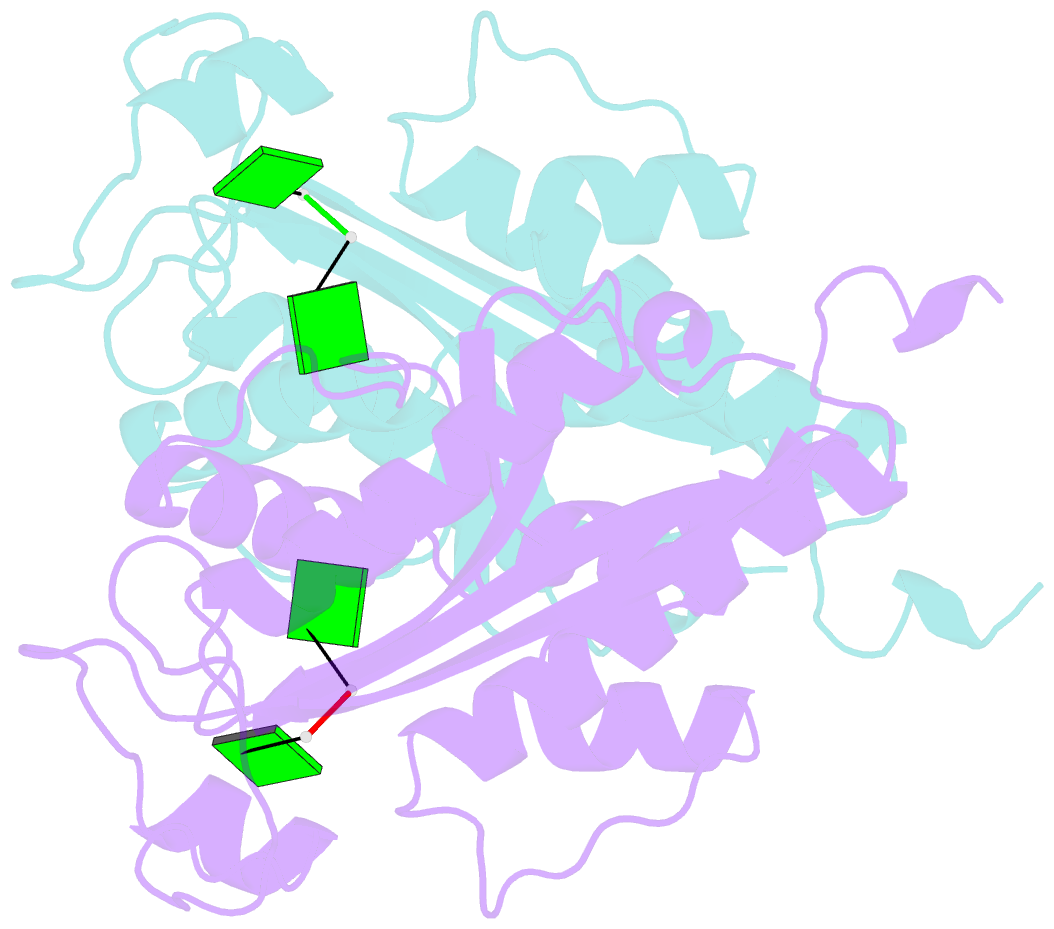
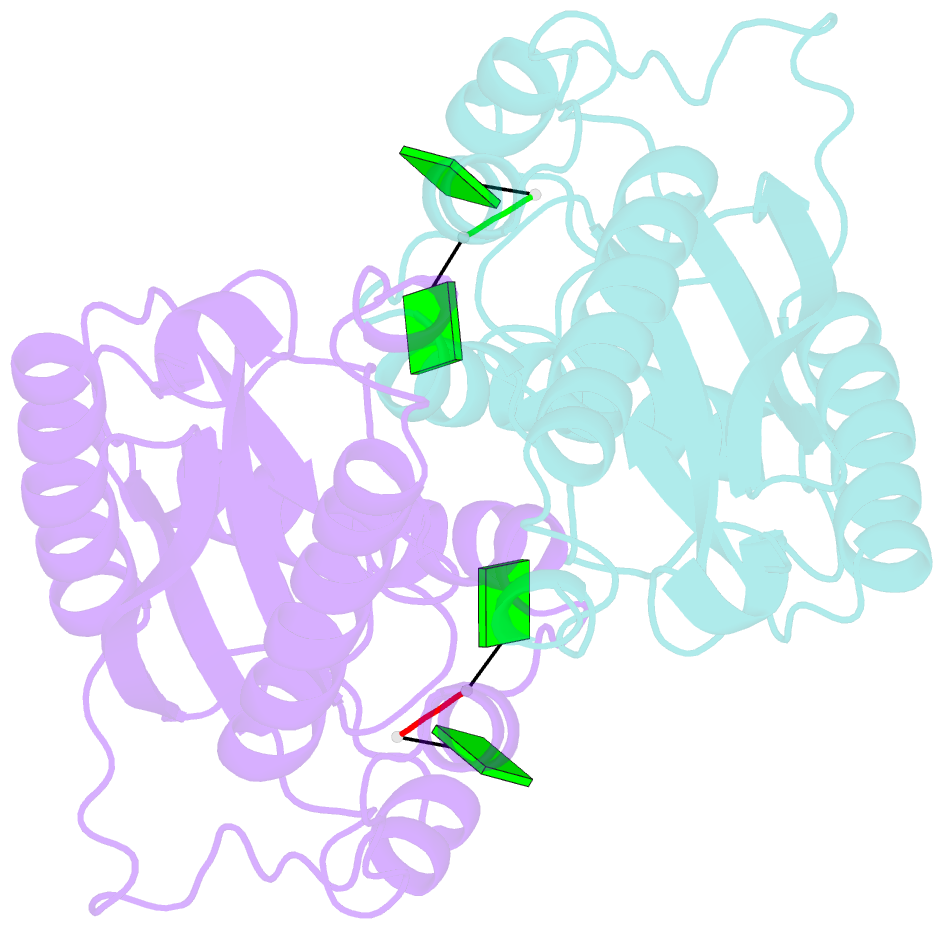
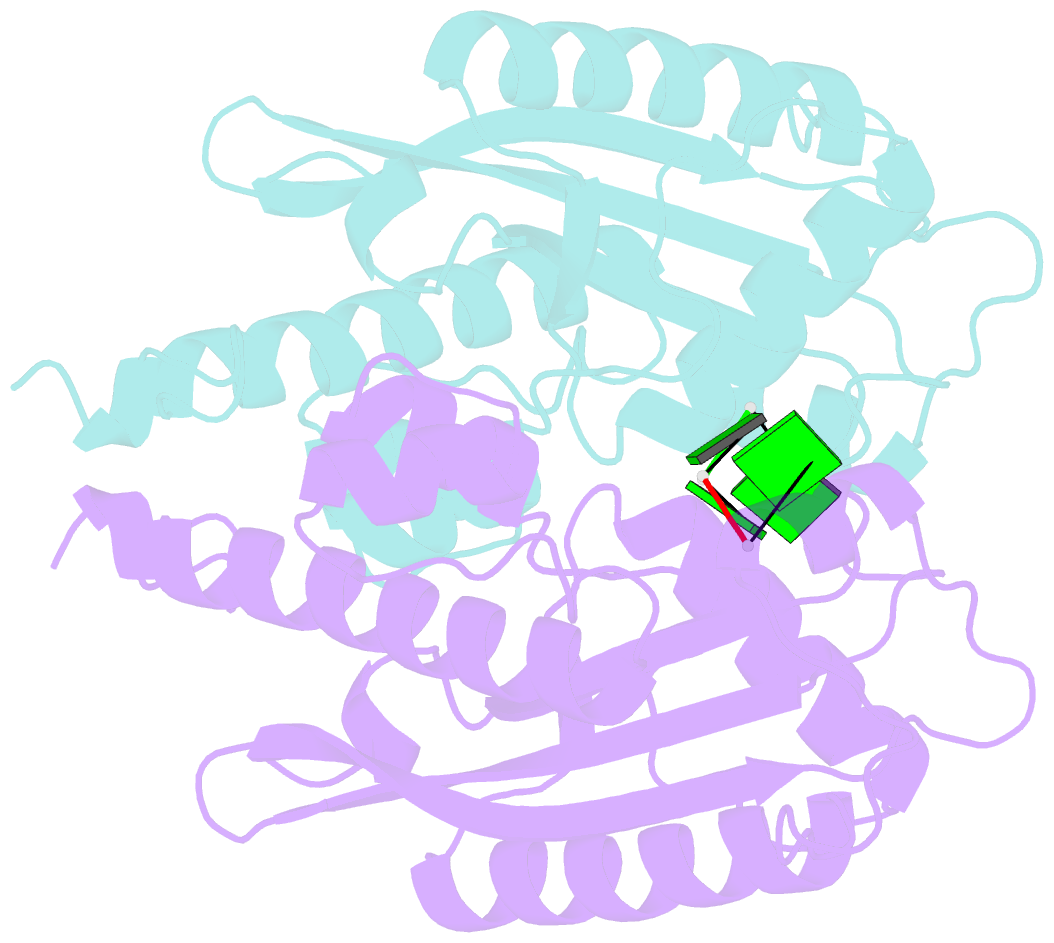
- Crystal structure of rnase t in complex with a di-nucleotide product (gg) with one mg in the active site
- Reference
- Hsiao Y-Y, Duh Y, Chen YP, Wang YT, Yuan HS (2012): "How an exonuclease decides where to stop in trimming of nucleic acids: crystal structures of RNase T-product complexes." Nucleic Acids Res., 40, 8144-8154. doi: 10.1093/nar/gks548.
- Abstract
- Exonucleases are key enzymes in the maintenance of genome stability, processing of immature RNA precursors and degradation of unnecessary nucleic acids. However, it remains unclear how exonucleases digest nucleic acids to generate correct end products for next-step processing. Here we show how the exonuclease RNase T stops its trimming precisely. The crystal structures of RNase T in complex with a stem-loop DNA, a GG dinucleotide and single-stranded DNA with different 3'-end sequences demonstrate why a duplex with a short 3'-overhang, a dinucleotide and a ssDNA with a 3'-end C cannot be further digested by RNase T. Several hydrophobic residues in RNase T change their conformation upon substrate binding and induce an active or inactive conformation in the active site that construct a precise machine to determine which substrate should be digested based on its sequence, length and structure. These studies thus provide mechanistic insights into how RNase T prevents over digestion of its various substrates, and the results can be extrapolated to the thousands of members of the DEDDh family of exonucleases.