Summary information and primary citation
- PDB-id
- 3vjr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.4 Å)
- Summary
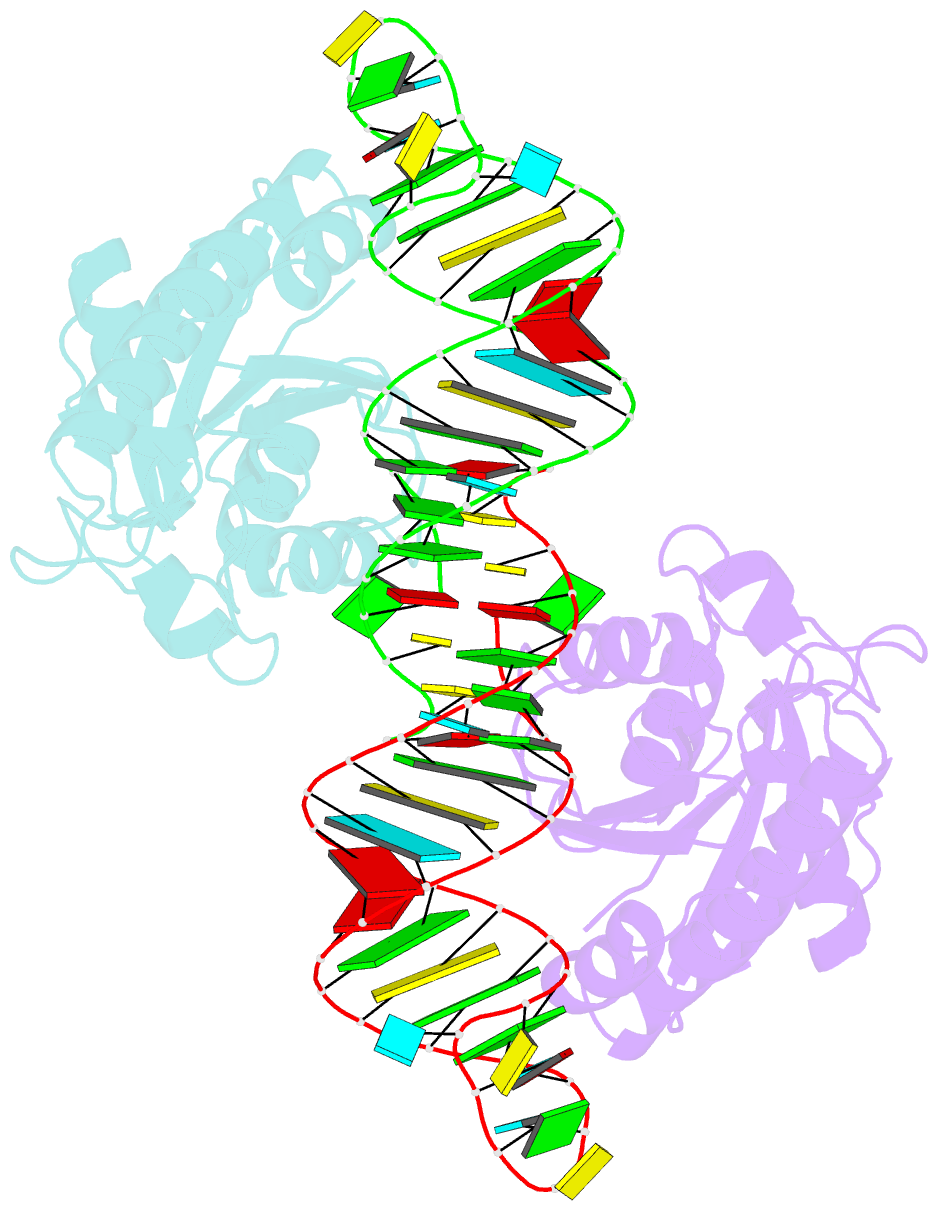
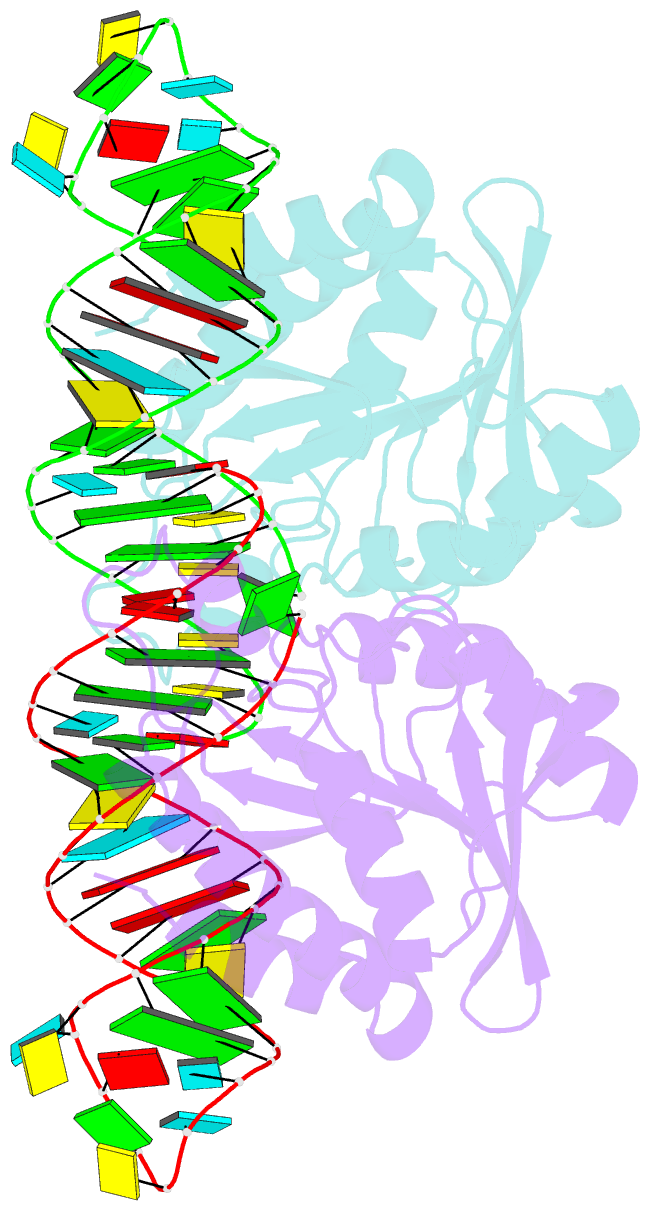
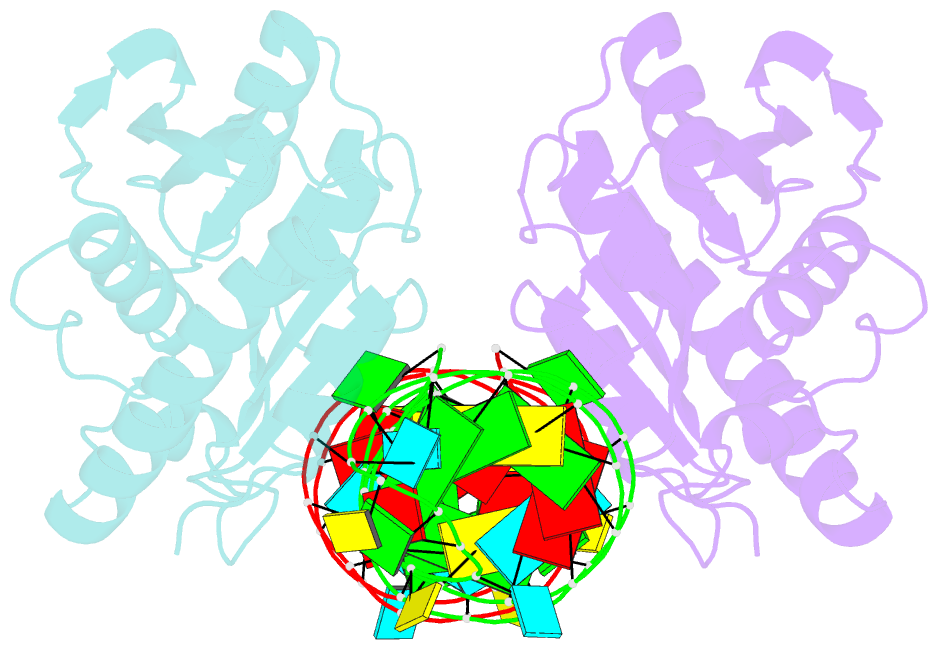
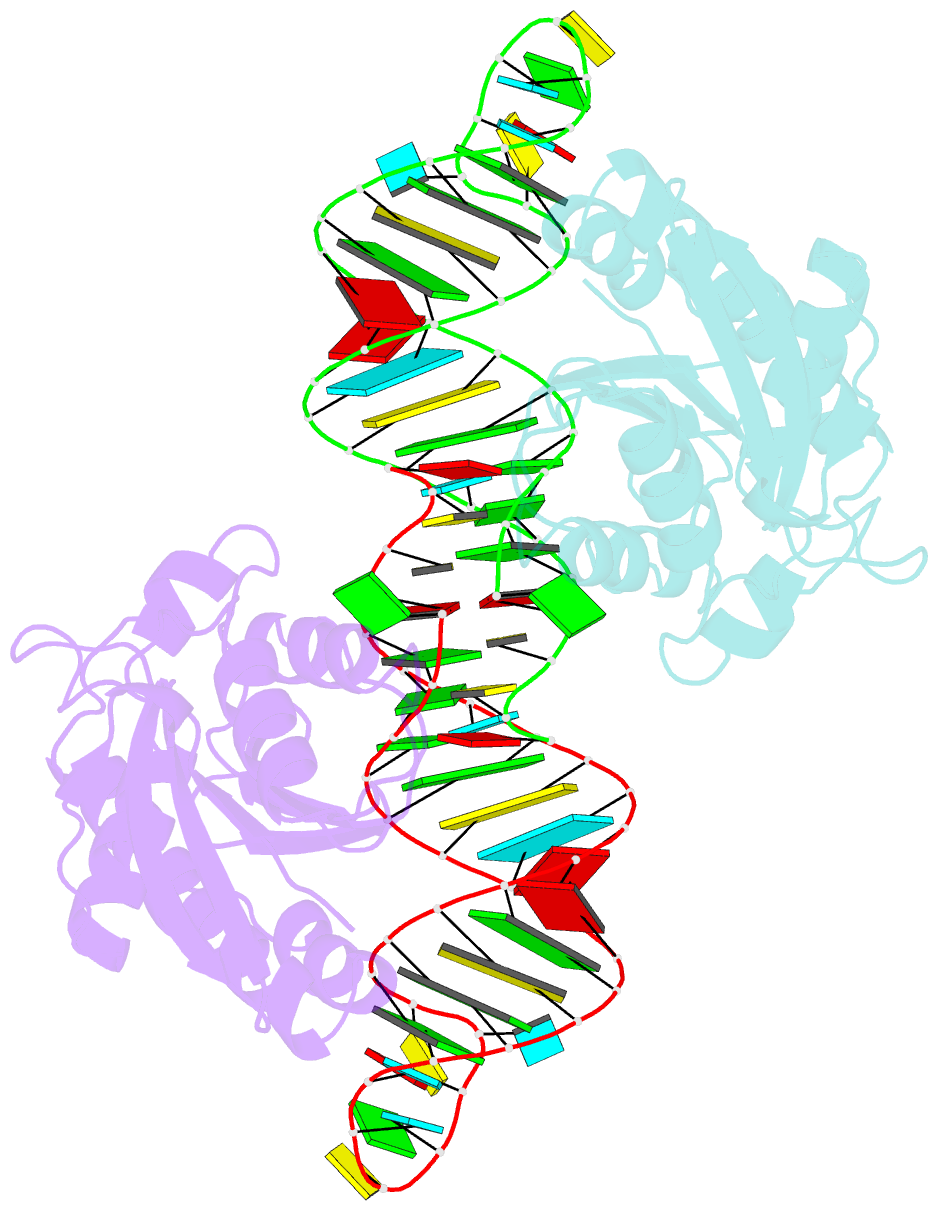
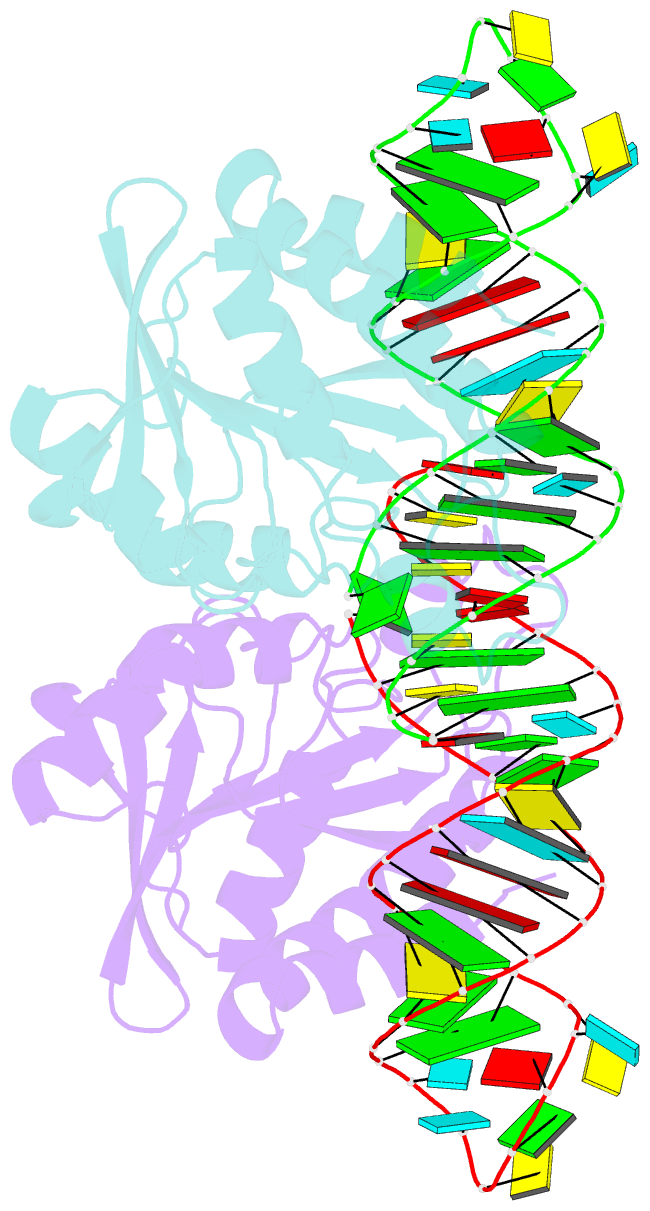
- Crystal structure of peptidyl-trna hydrolase from escherichia coli in complex with the cca-acceptor-t[psi]c domain of trna
- Reference
- Ito K, Murakami R, Mochizuki M, Qi H, Shimizu Y, Miura K, Ueda T, Uchiumi T (2012): "Structural basis for the substrate recognition and catalysis of peptidyl-tRNA hydrolase." Nucleic Acids Res., 40, 10521-10531. doi: 10.1093/nar/gks790.
- Abstract
- Peptidyl-tRNA hydrolase (Pth) cleaves the ester bond between the peptide and the tRNA of peptidyl-tRNA molecules, which are produced by aborted translation, to recycle tRNA for further rounds of protein synthesis. Pth is ubiquitous in nature, and its enzymatic activity is essential for bacterial viability. We have determined the crystal structure of Escherichia coli Pth in complex with the tRNA CCA-acceptor-TΨC domain, the enzyme-binding region of the tRNA moiety of the substrate, at 2.4 Å resolution. In combination with site-directed mutagenesis studies, the structure identified the amino acid residues involved in tRNA recognition. The structure also revealed that Pth interacts with the tRNA moiety through the backbone phosphates and riboses, and no base-specific interactions were observed, except for the interaction with the highly conserved base G53. This feature enables Pth to accept the diverse sequences of the elongator-tRNAs as substrate components. Furthermore, we propose an authentic Pth:peptidyl-tRNA complex model and a detailed mechanism for the hydrolysis reaction, based on the present crystal structure and the previous studies' results.