Summary information and primary citation
- PDB-id
- 3vnu; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation, transferase-RNA
- Method
- X-ray (3.2 Å)
- Summary
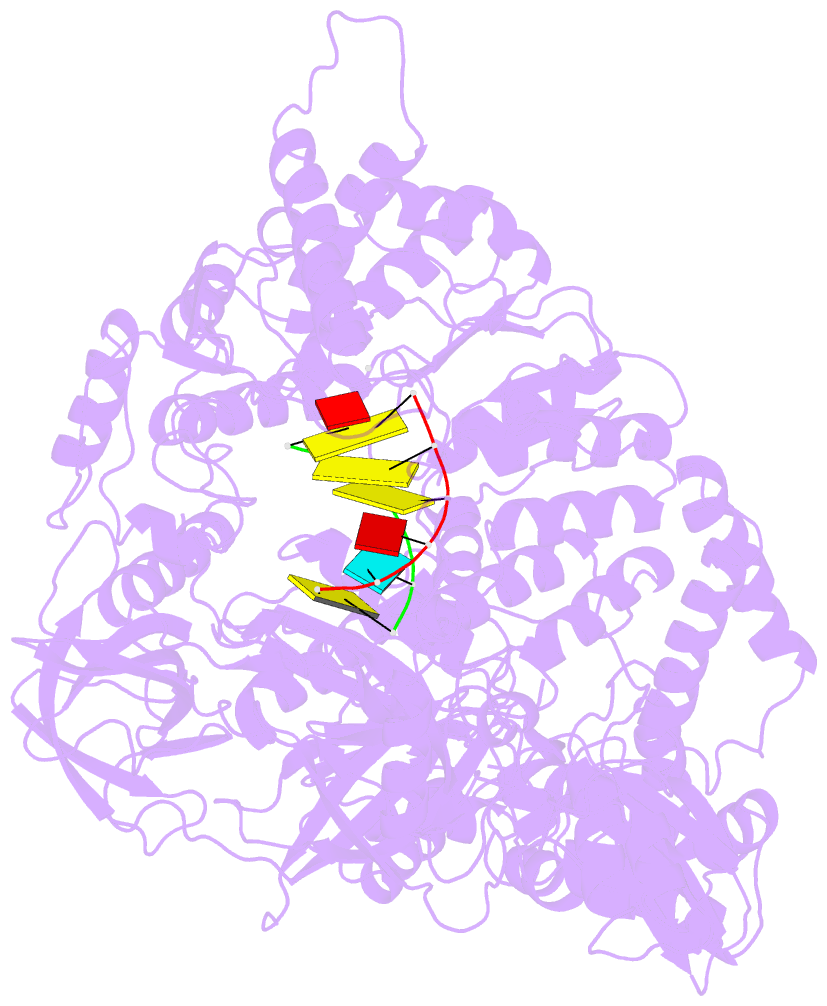
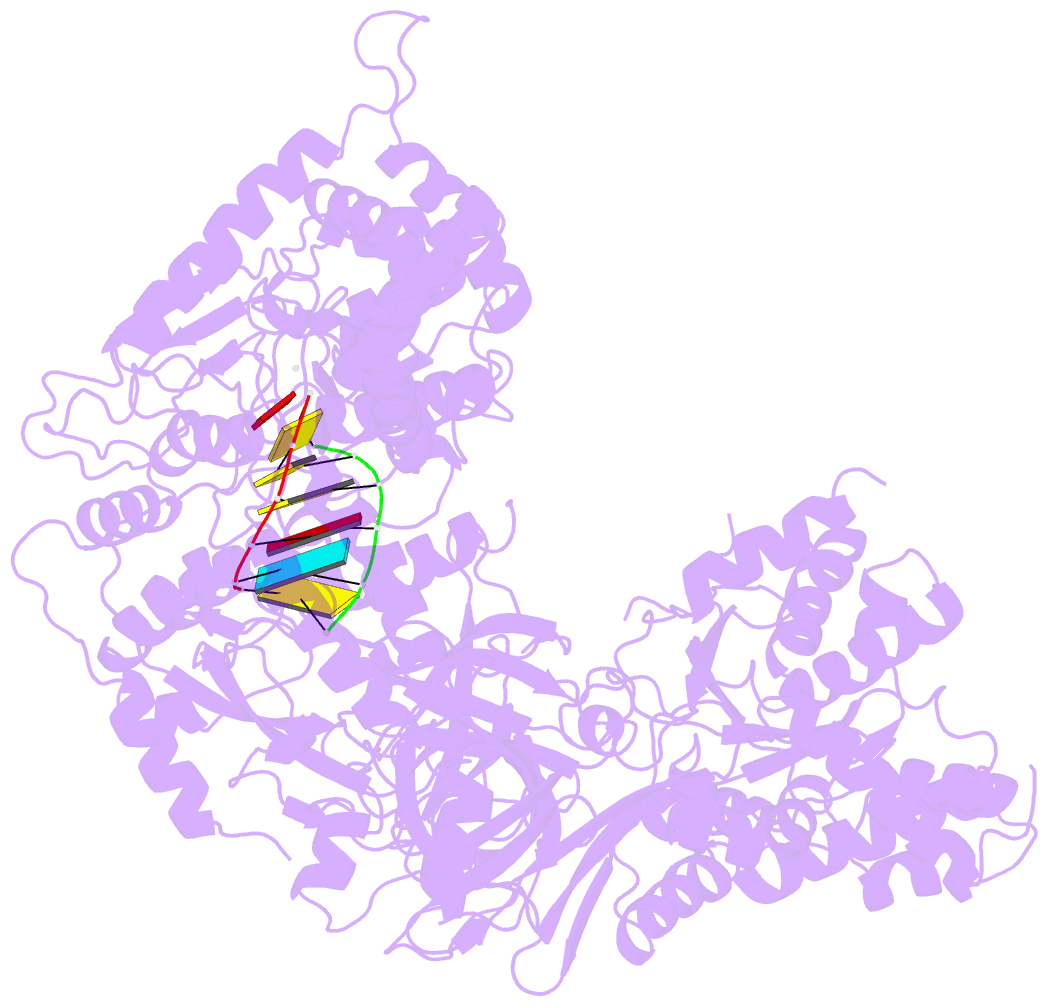
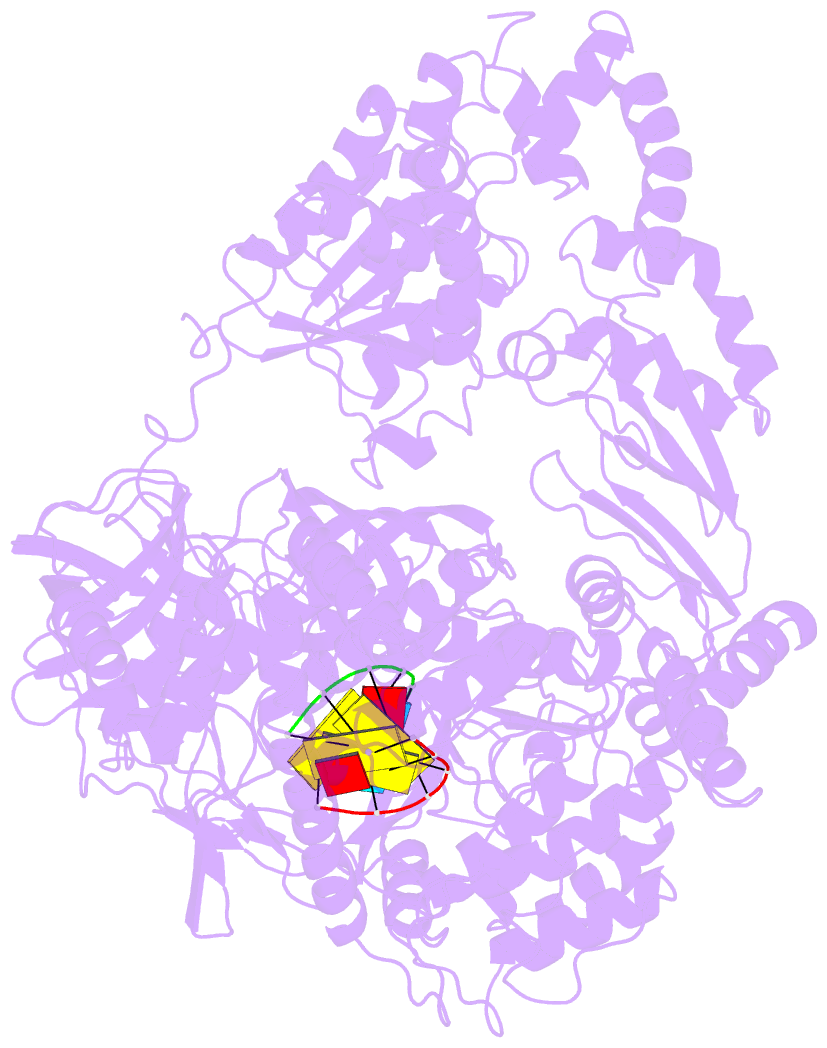
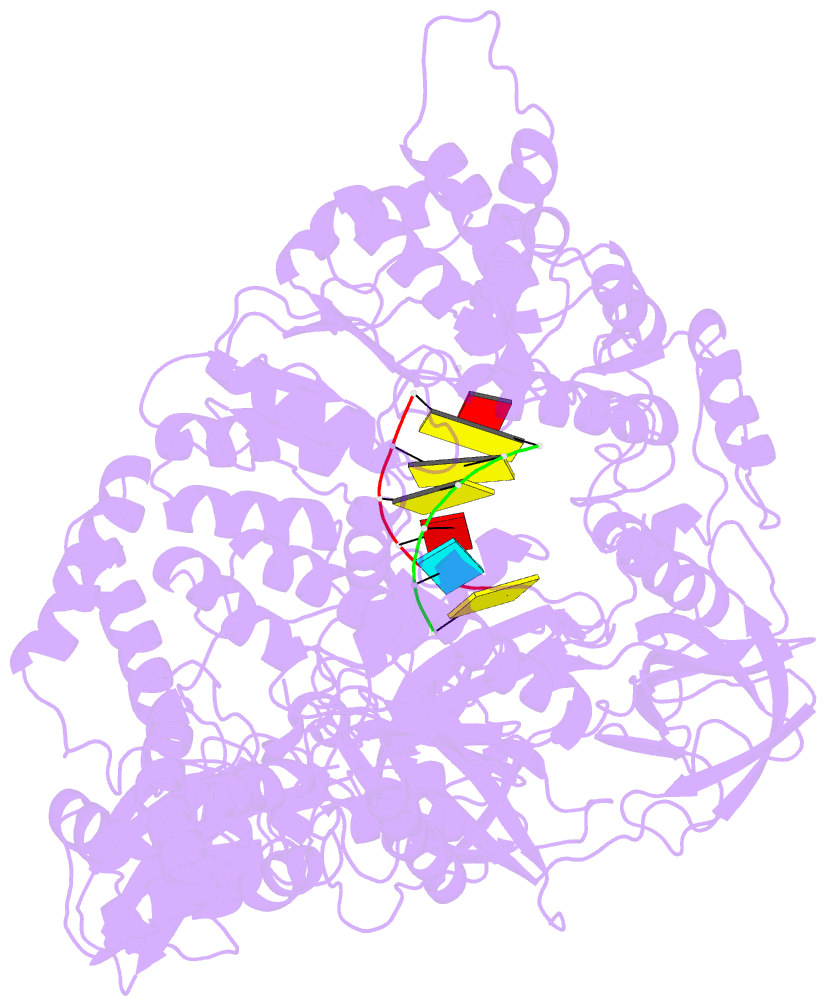
- Complex structure of viral RNA polymerase i
- Reference
- Takeshita D, Yamashita S, Tomita K (2012): "Mechanism for template-independent terminal adenylation activity of Q beta replicase." Structure, 20, 1661-1669. doi: 10.1016/j.str.2012.07.004.
- Abstract
- The genomic RNA of Qβ virus is replicated by Qβ replicase, a template-dependent RNA polymerase complex. Qβ replicase has an intrinsic template-independent RNA 3'-adenylation activity, which is required for efficient viral RNA amplification in the host cells. However, the mechanism of the template-independent 3'-adenylation of RNAs by Qβ replicase has remained elusive. We determined the structure of a complex that includes Qβ replicase, a template RNA, a growing RNA complementary to the template RNA, and ATP. The structure represents the terminal stage of RNA polymerization and reveals that the shape and size of the nucleotide-binding pocket becomes available for ATP accommodation after the 3'-penultimate template-dependent C-addition. The stacking interaction between the ATP and the neighboring Watson-Crick base pair, between the 5'-G in the template and the 3'-C in the growing RNA, contributes to the nucleotide specificity. Thus, the template for the template-independent 3'-adenylation by Qβ replicase is the RNA and protein ribonucleoprotein complex.