Summary information and primary citation
- PDB-id
- 3vw3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-DNA
- Method
- X-ray (2.5 Å)
- Summary
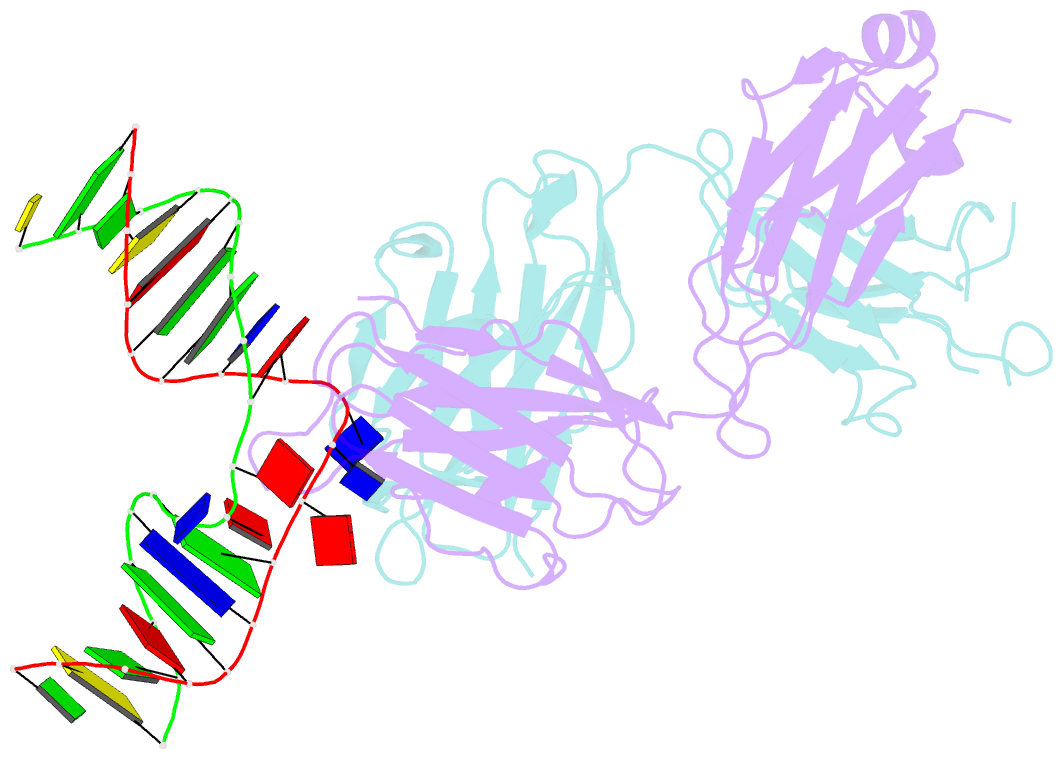
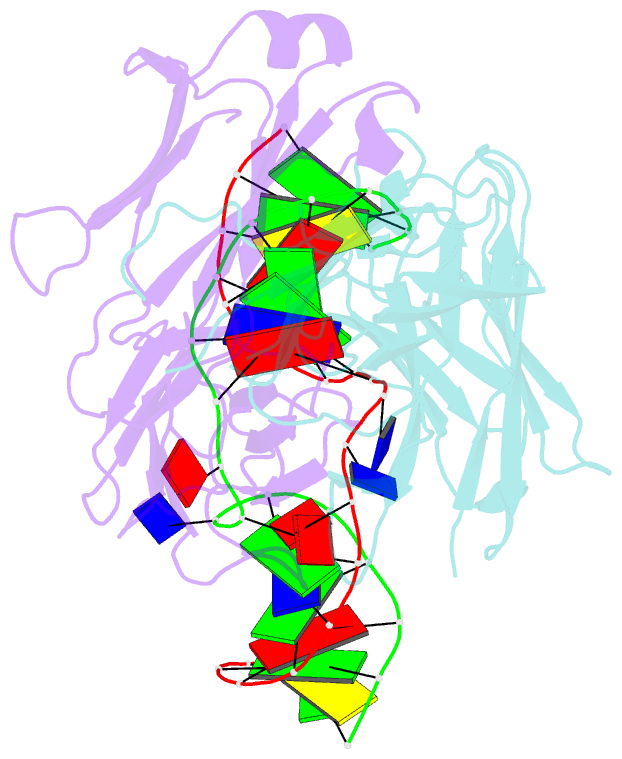
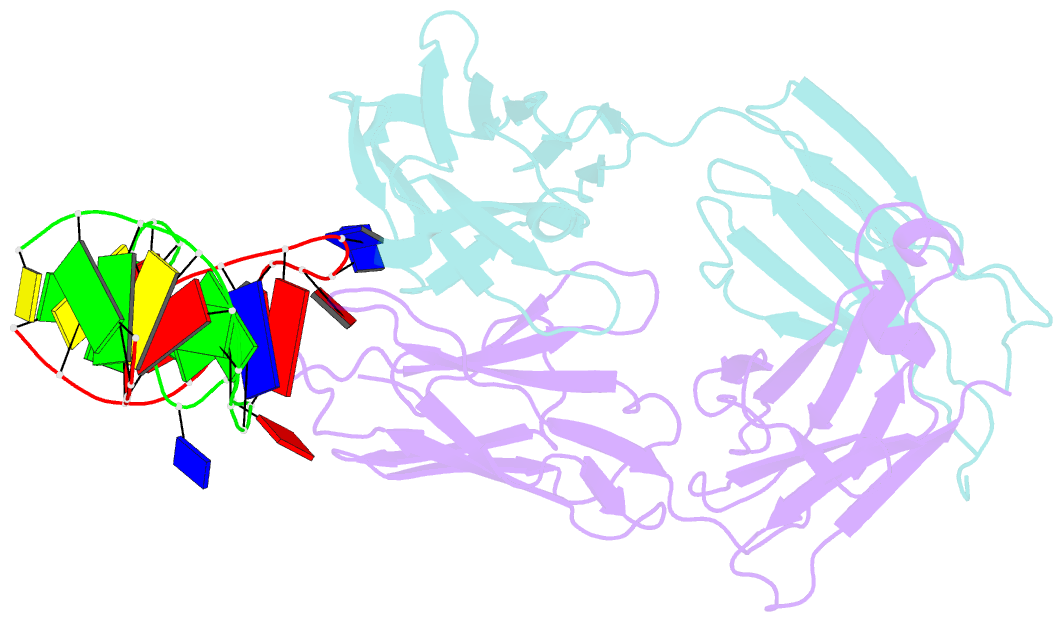
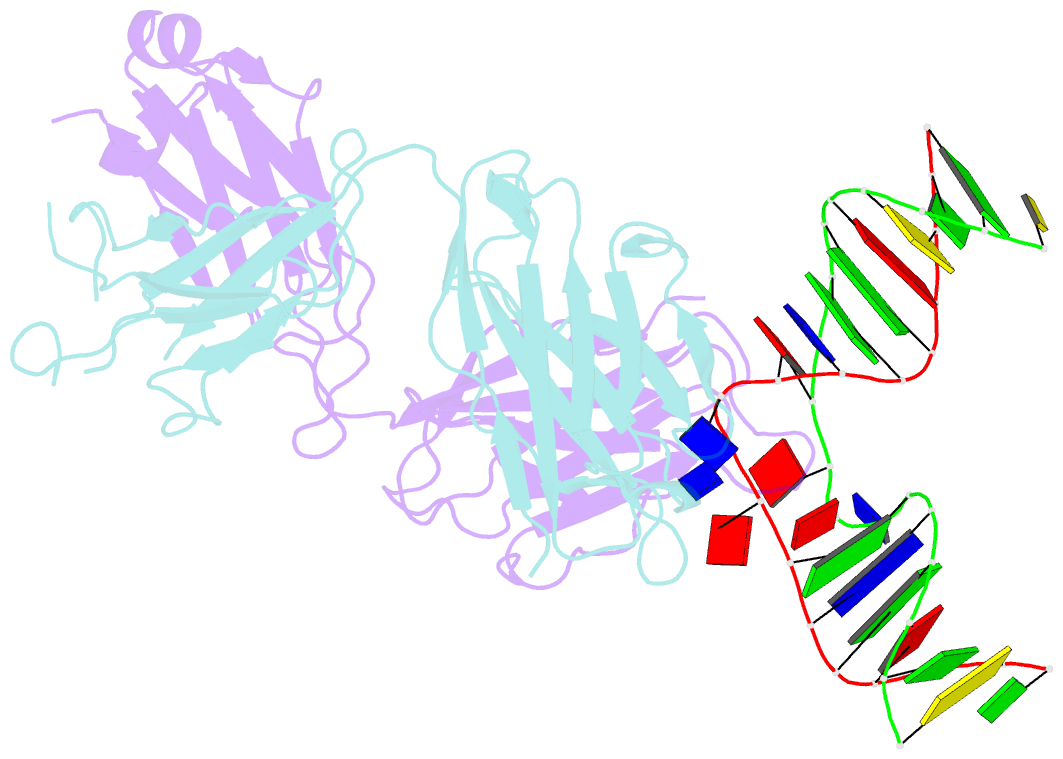
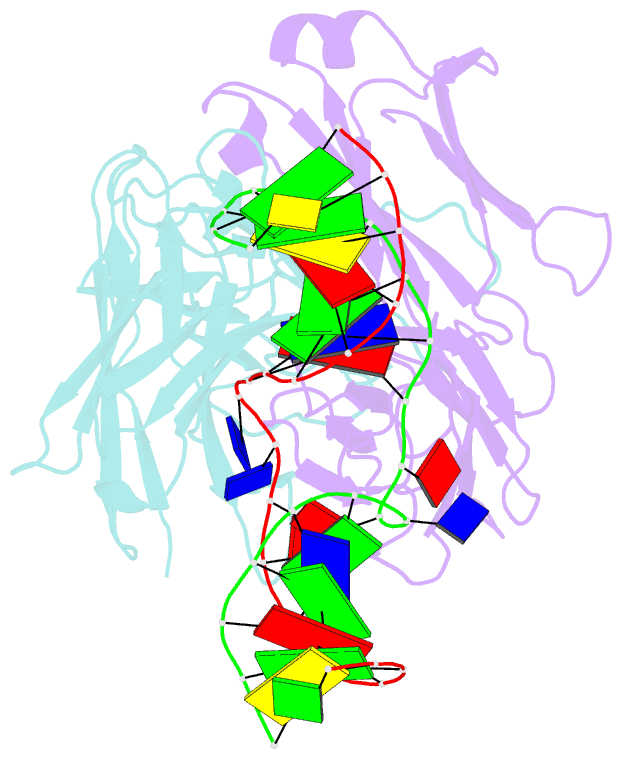
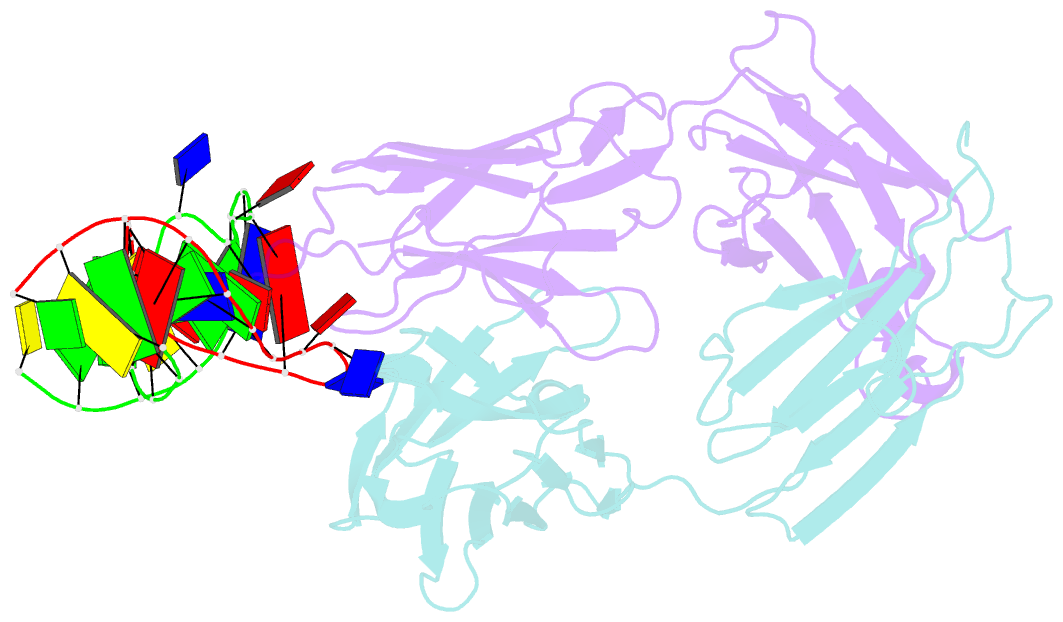
- Antibody 64m-5 fab in complex with a double-stranded DNA (6-4) photoproduct
- Reference
- Yokoyama H, Mizutani R, Satow Y (2013): "Structure of a double-stranded DNA (6-4) photoproduct in complex with the 64M-5 antibody Fab." Acta Crystallogr.,Sect.D, 69, 504-512. doi: 10.1107/S0907444912050007.
- Abstract
- DNA photoproducts with (6-4) pyrimidine-pyrimidone adducts formed by ultraviolet radiation have been implicated in mutagenesis and cancer. The crystal structure of double-stranded DNA containing the (6-4) photoproduct in complex with the anti-(6-4)-photoproduct antibody 64M-5 Fab was determined at 2.5 Å resolution. The T(6-4)T segment and the 5'-side adjacent adenosine are flipped out of the duplex and are accommodated in the concave antigen-binding pocket composed of six complementarity-determining regions (CDRs). A loop comprised of CDR L1 residues is inserted between the flipped-out T(6-4)T segment and the complementary DNA. The separation of strands by the insertion of the loop facilitates extensive and specific recognition of the photoproduct. The DNA helices flanking the T(6-4)T segment are kinked by 87°. The 64M-5 Fab recognizes the T(6-4)T segment dissociated from the complementary strand, indicating that the (6-4) photoproduct can be detected in double-stranded DNA as well as in single-stranded DNA using the 64M-5 antibody. The structure and recognition mode of the 64M-5 antibody were compared with those of the DNA (6-4) photolyase and nucleotide-excision repair protein DDB1-DDB2. These proteins have distinctive binding-site structures that are appropriate for their functions, and the flipping out of the photolesion and the kinking of the DNA are common to mutagenic (6-4) photoproducts recognized by proteins.