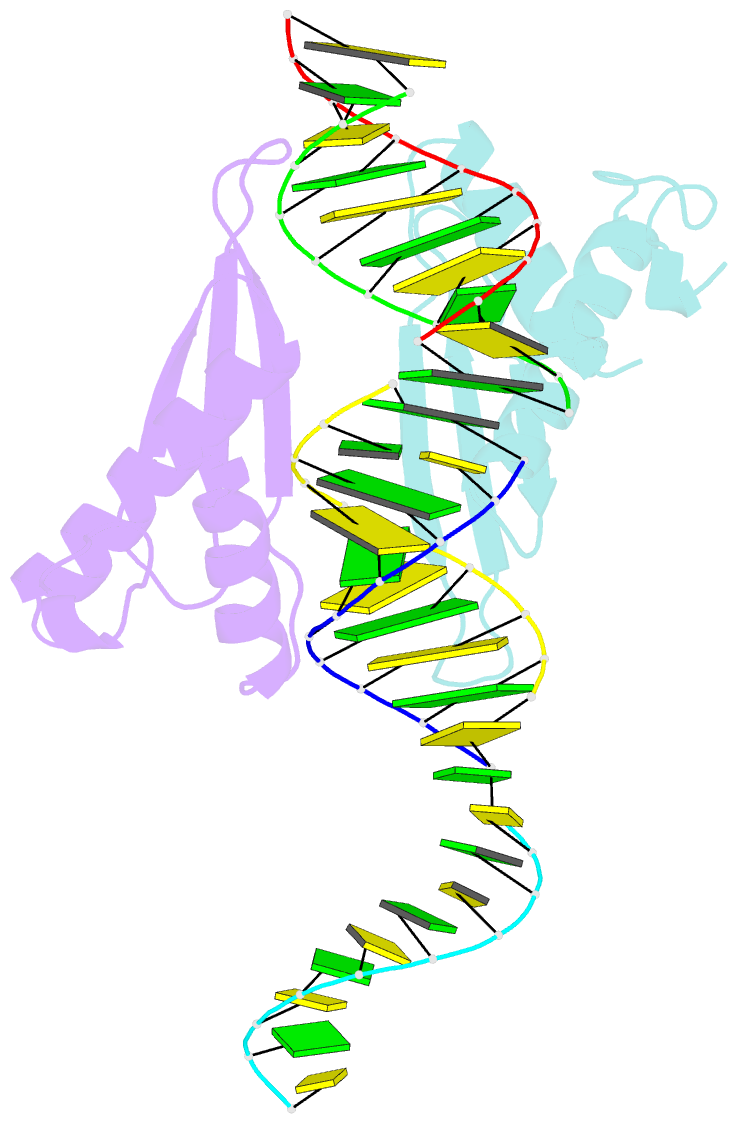
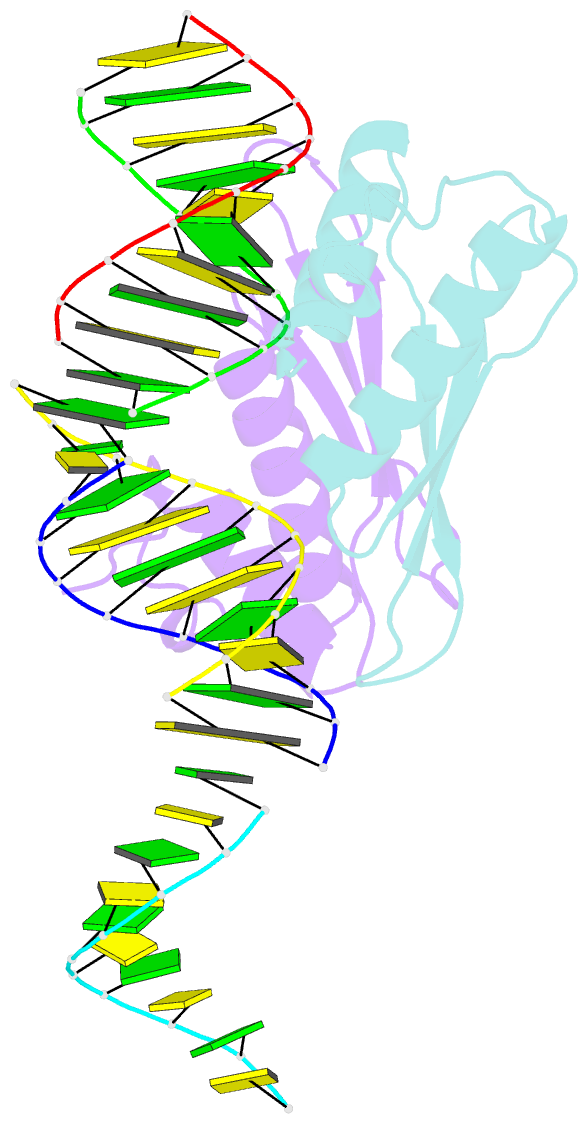
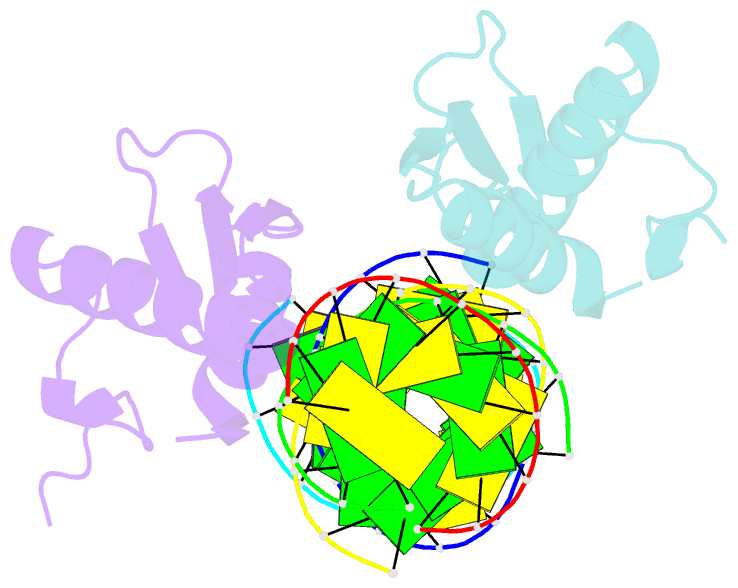
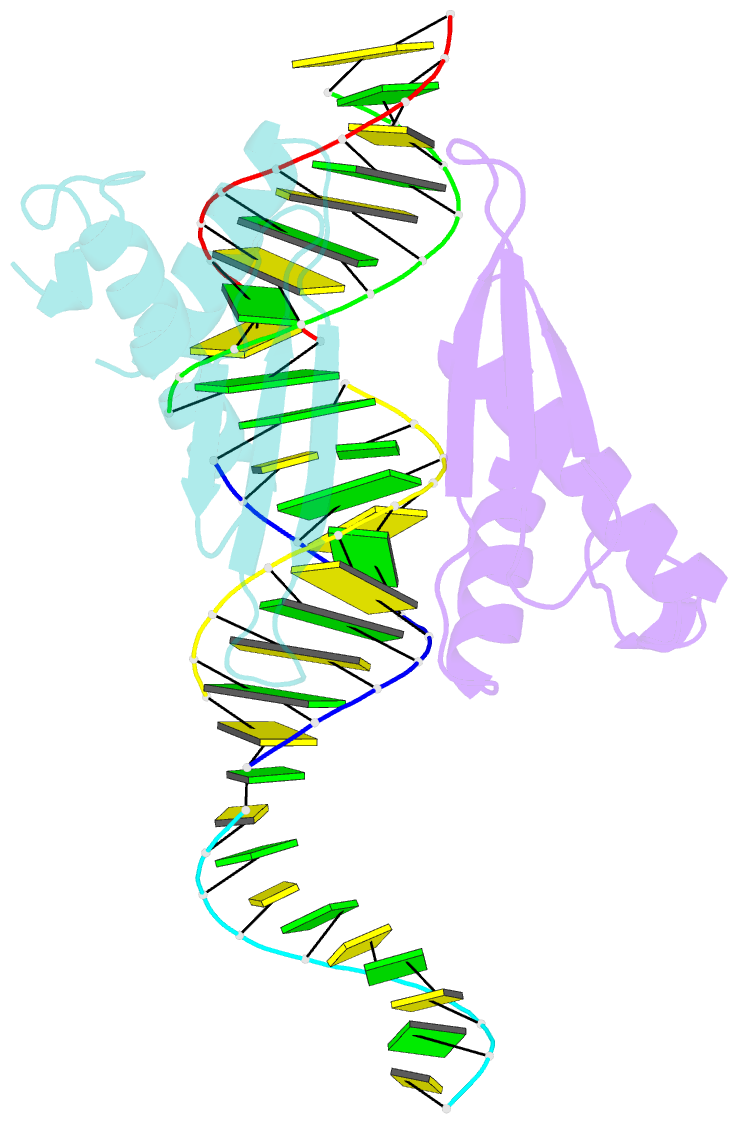
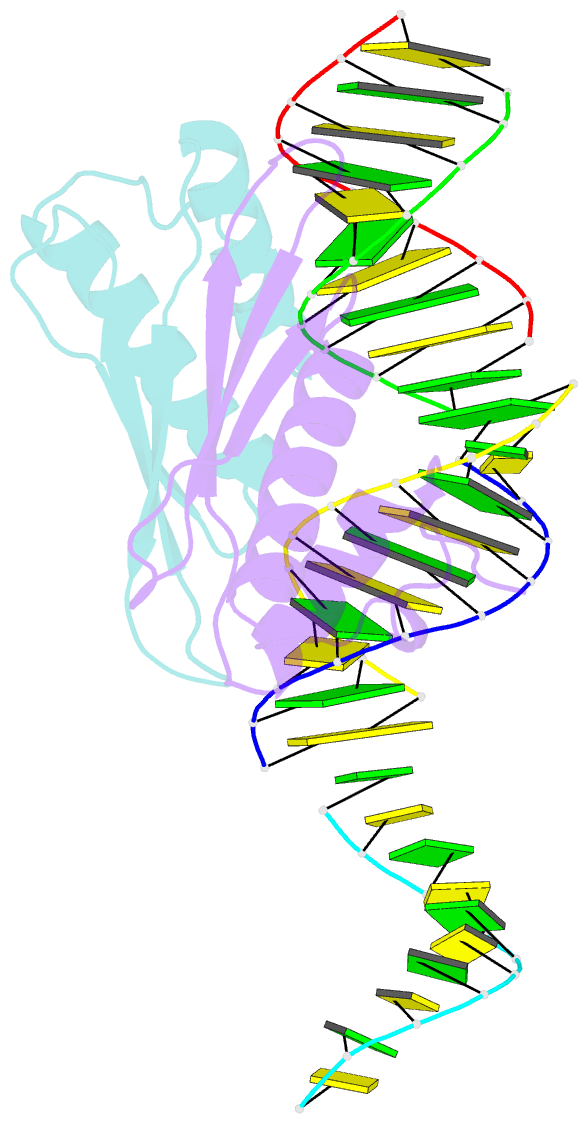
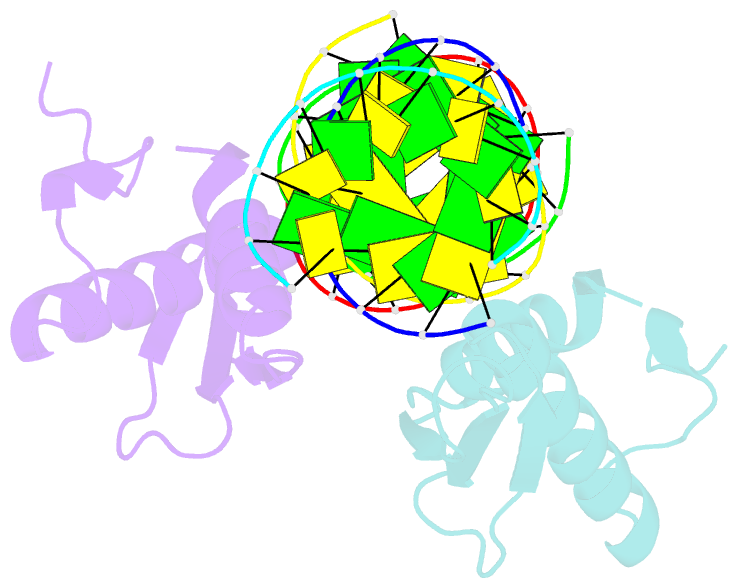
Summary information and primary citation
- PDB-id
- 3vyy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.9 Å)
- Summary
- Structural insights into risc assembly facilitated by dsrna binding domains of human RNA helicase a (dhx9)
- Reference
- Fu Q, Yuan YA (2013): "Structural insights into RISC assembly facilitated by dsRNA-binding domains of human RNA helicase A (DHX9)." Nucleic Acids Res., 41, 3457-3470. doi: 10.1093/nar/gkt042.
- Abstract
- Intensive research interest has focused on small RNA-processing machinery and the RNA-induced silencing complex (RISC), key cellular machines in RNAi pathways. However, the structural mechanism regarding RISC assembly, the primary step linking small RNA processing and RNA-mediated gene silencing, is largely unknown. Human RNA helicase A (DHX9) was reported to function as an RISC-loading factor, and such function is mediated mainly by its dsRNA-binding domains (dsRBDs). Here, we report the crystal structures of human RNA helicase A (RHA) dsRBD1 and dsRBD2 domains in complex with dsRNAs, respectively. Structural analysis not only reveals higher siRNA duplex-binding affinity displayed by dsRBD1, but also identifies a crystallographic dsRBD1 pair of physiological significance in cooperatively recognizing dsRNAs. Structural observations are further validated by isothermal titration calorimetric (ITC) assay. Moreover, co-immunoprecipitation (co-IP) assay coupled with mutagenesis demonstrated that both dsRBDs are required for RISC association, and such association is mediated by dsRNA. Hence, our structural and functional efforts have revealed a potential working model for siRNA recognition by RHA tandem dsRBDs, and together they provide direct structural insights into RISC assembly facilitated by RHA.