Summary information and primary citation
- PDB-id
- 3wpg; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.246 Å)
- Summary
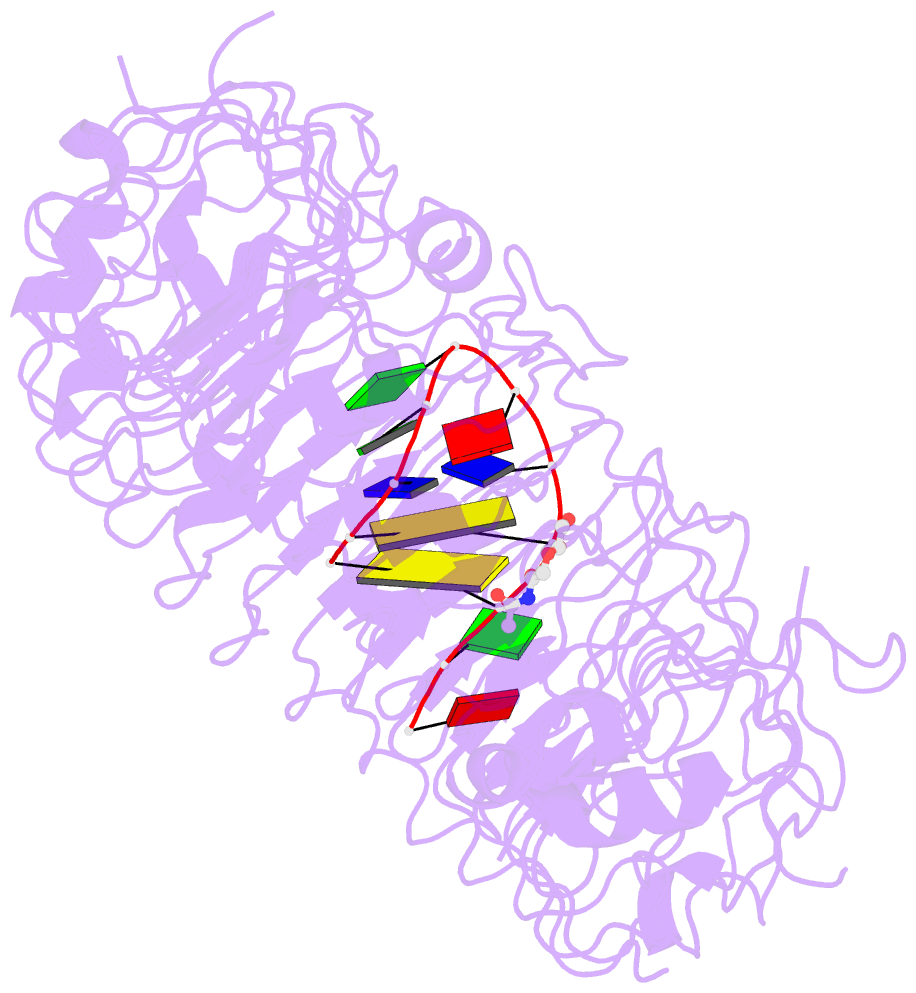
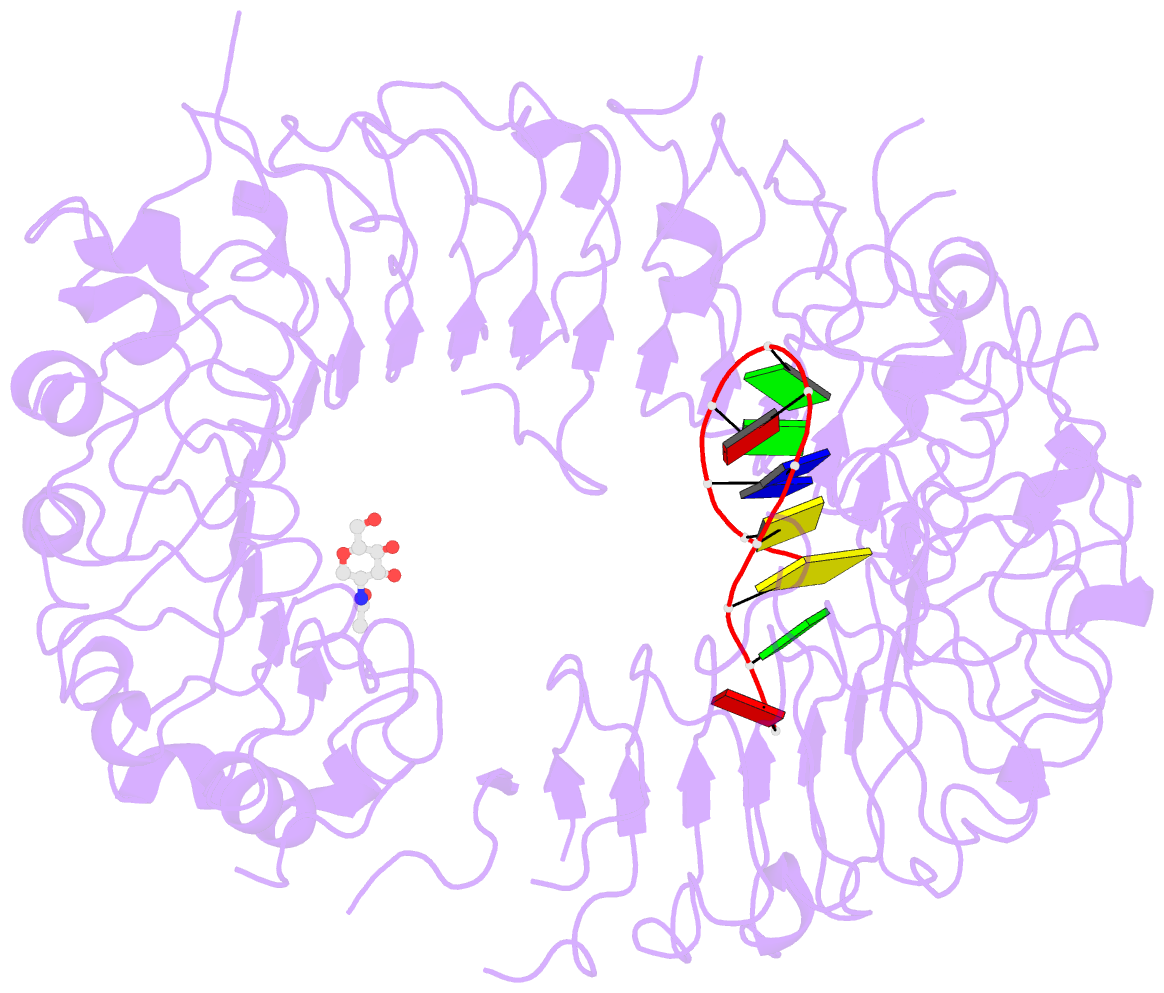
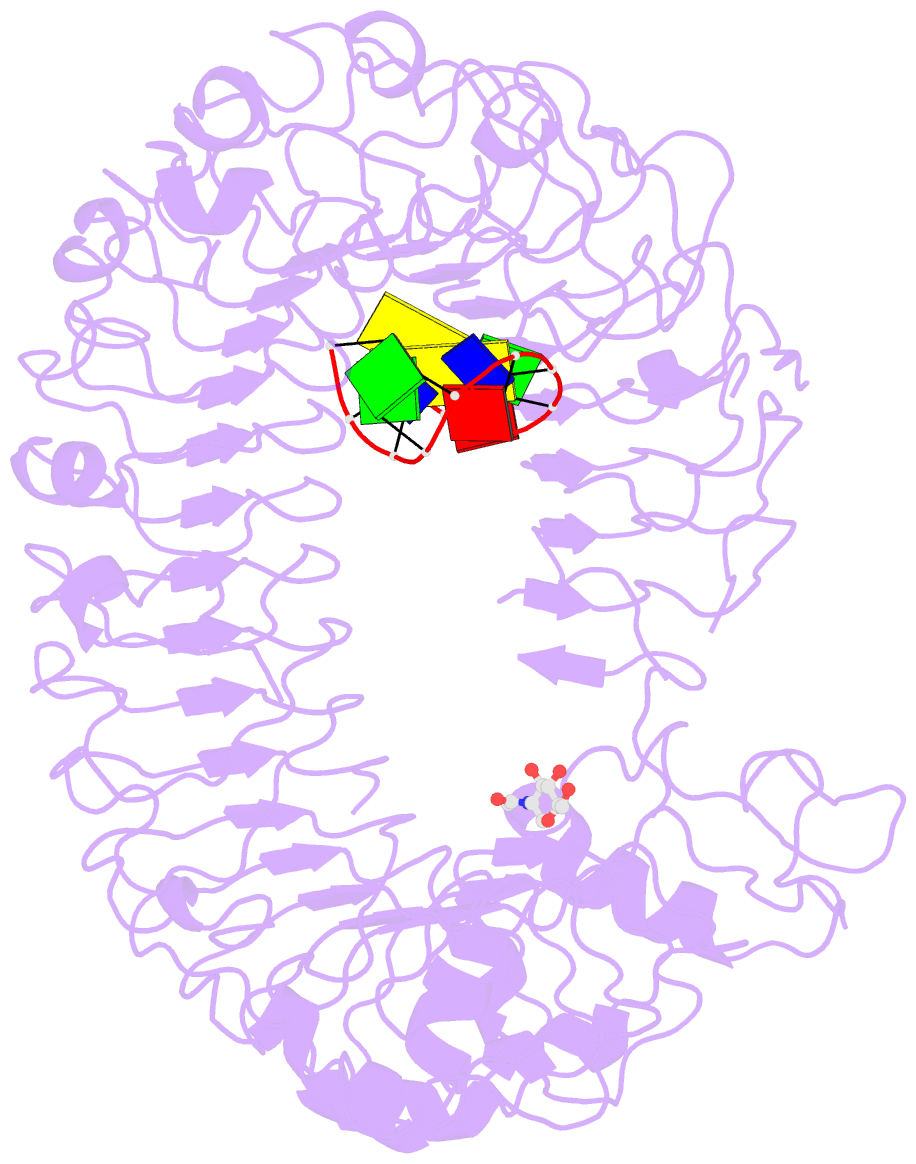
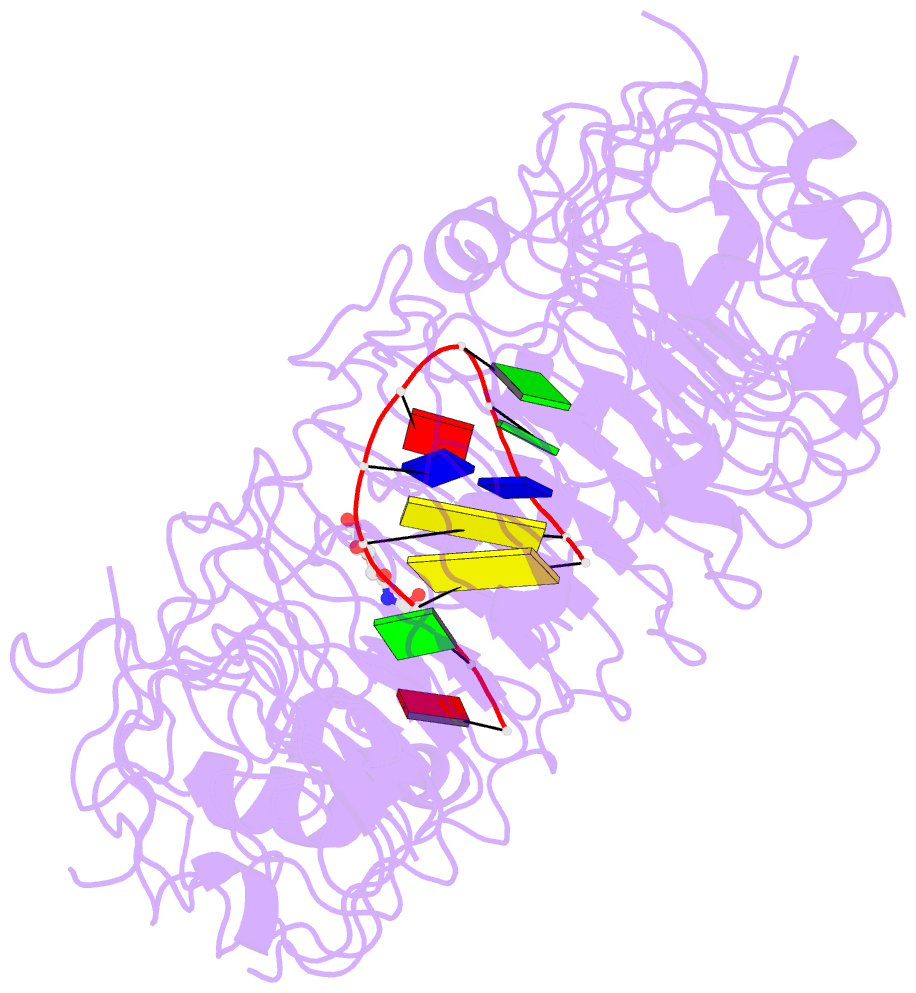
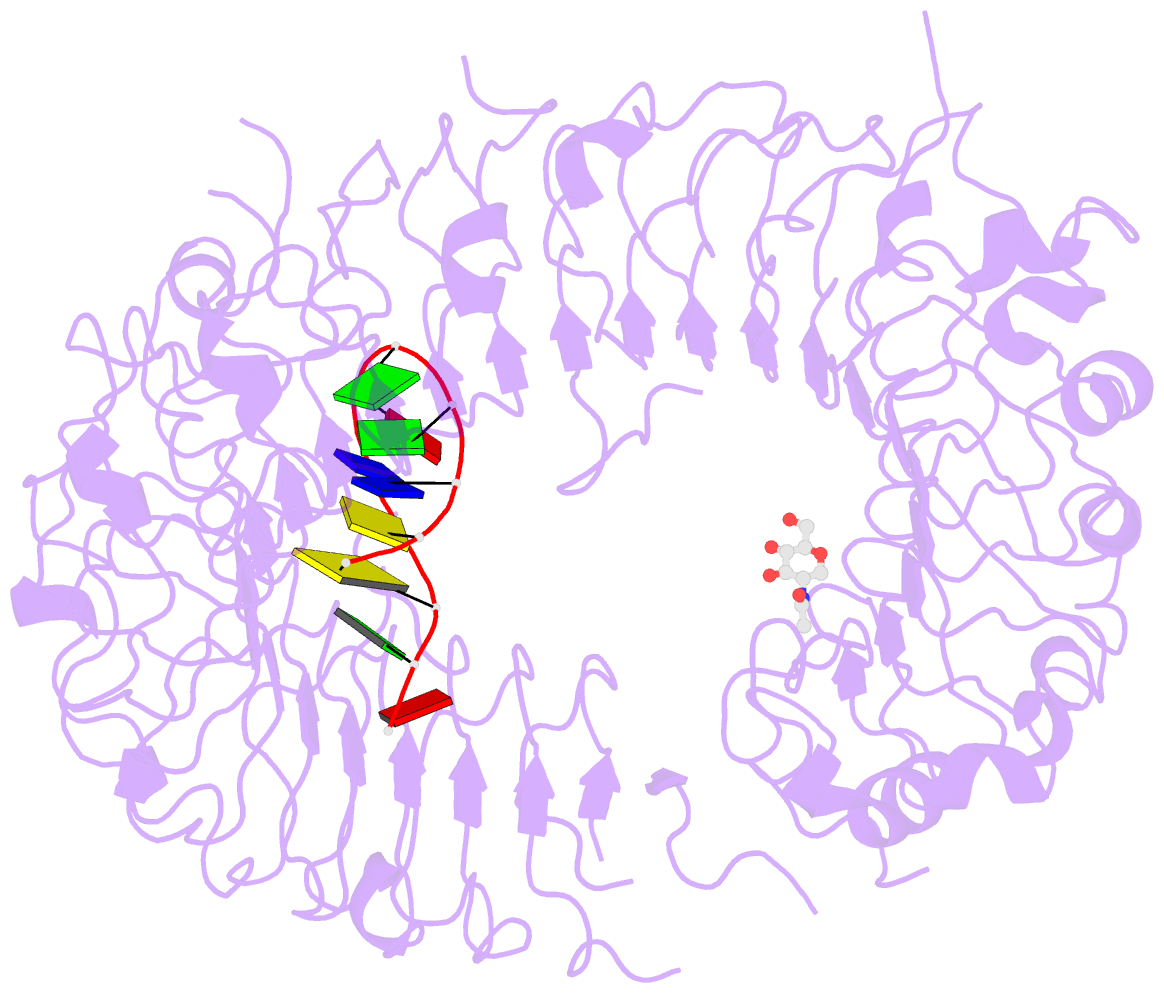
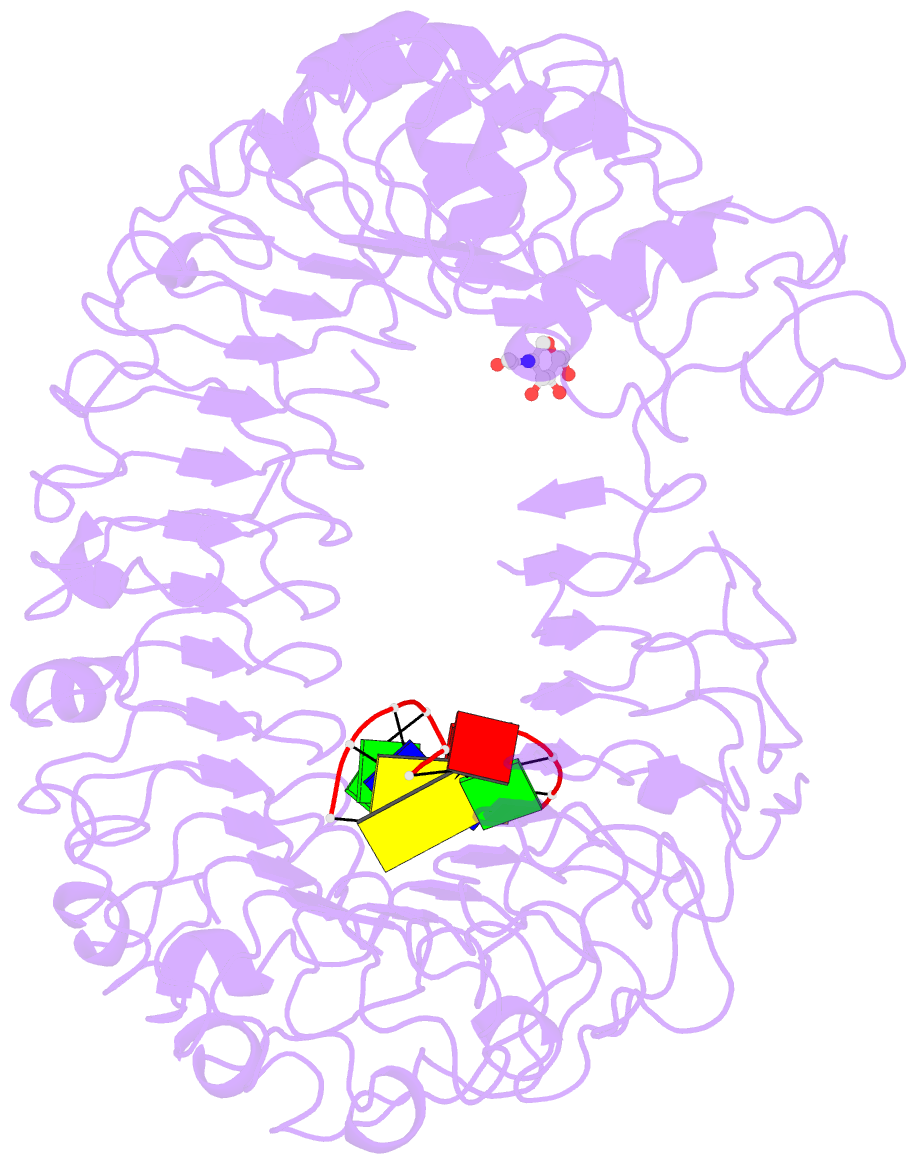
- Crystal structure of mouse tlr9 in complex with inhibitory dna4084 (form 1)
- Reference
- Ohto U, Shibata T, Tanji H, Ishida H, Krayukhina E, Uchiyama S, Miyake K, Shimizu T (2015): "Structural basis of CpG and inhibitory DNA recognition by Toll-like receptor 9." Nature, 520, 702-705. doi: 10.1038/nature14138.
- Abstract
- Innate immunity serves as the first line of defence against invading pathogens such as bacteria and viruses. Toll-like receptors (TLRs) are examples of innate immune receptors, which sense specific molecular patterns from pathogens and activate immune responses. TLR9 recognizes bacterial and viral DNA containing the cytosine-phosphate-guanine (CpG) dideoxynucleotide motif. The molecular basis by which CpG-containing DNA (CpG-DNA) elicits immunostimulatory activity via TLR9 remains to be elucidated. Here we show the crystal structures of three forms of TLR9: unliganded, bound to agonistic CpG-DNA, and bound to inhibitory DNA (iDNA). Agonistic-CpG-DNA-bound TLR9 formed a symmetric TLR9-CpG-DNA complex with 2:2 stoichiometry, whereas iDNA-bound TLR9 was a monomer. CpG-DNA was recognized by both protomers in the dimer, in particular by the amino-terminal fragment (LRRNT-LRR10) from one protomer and the carboxy-terminal fragment (LRR20-LRR22) from the other. The iDNA, which formed a stem-loop structure suitable for binding by intramolecular base pairing, bound to the concave surface from LRR2-LRR10. This structure serves as an important basis for improving our understanding of the functional mechanisms of TLR9.