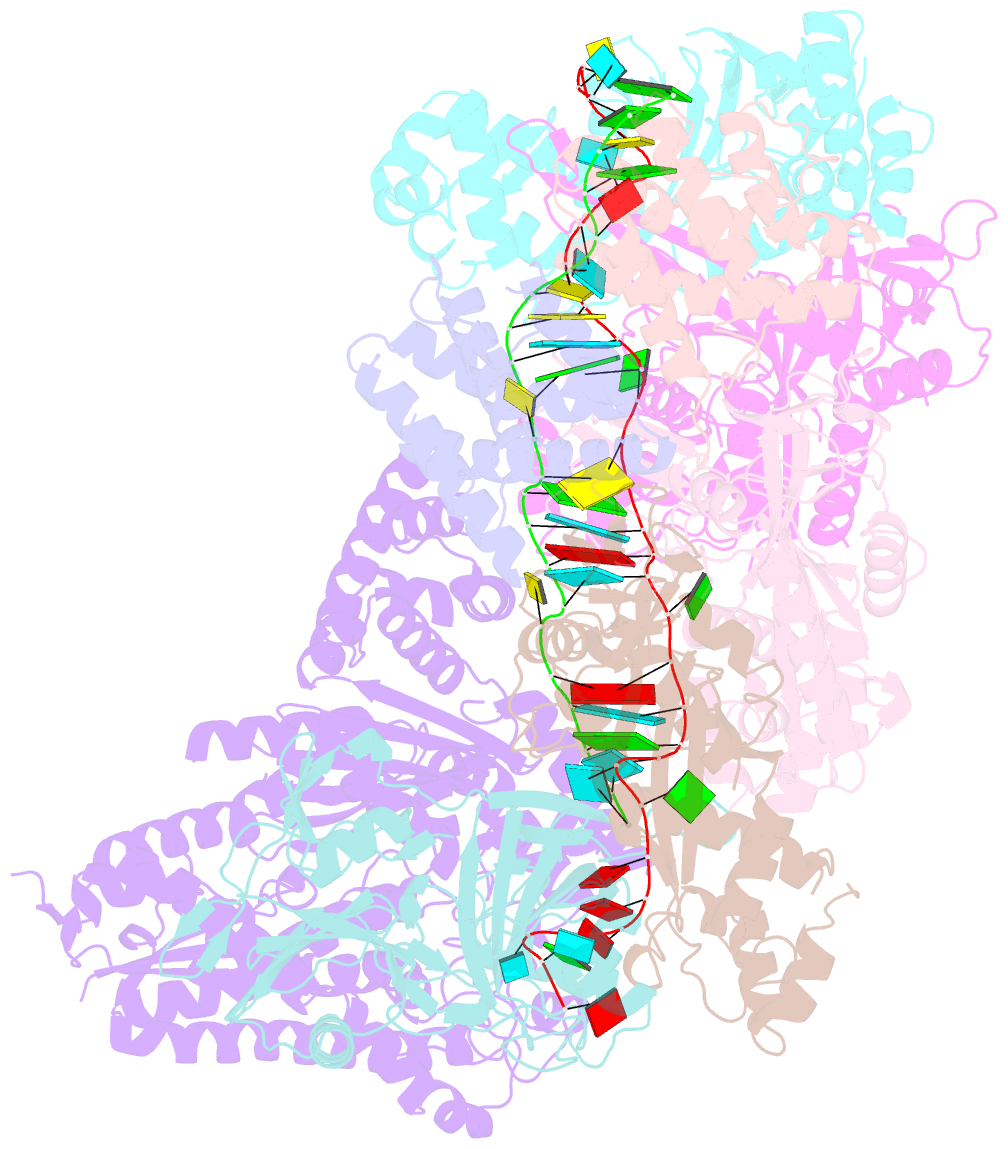
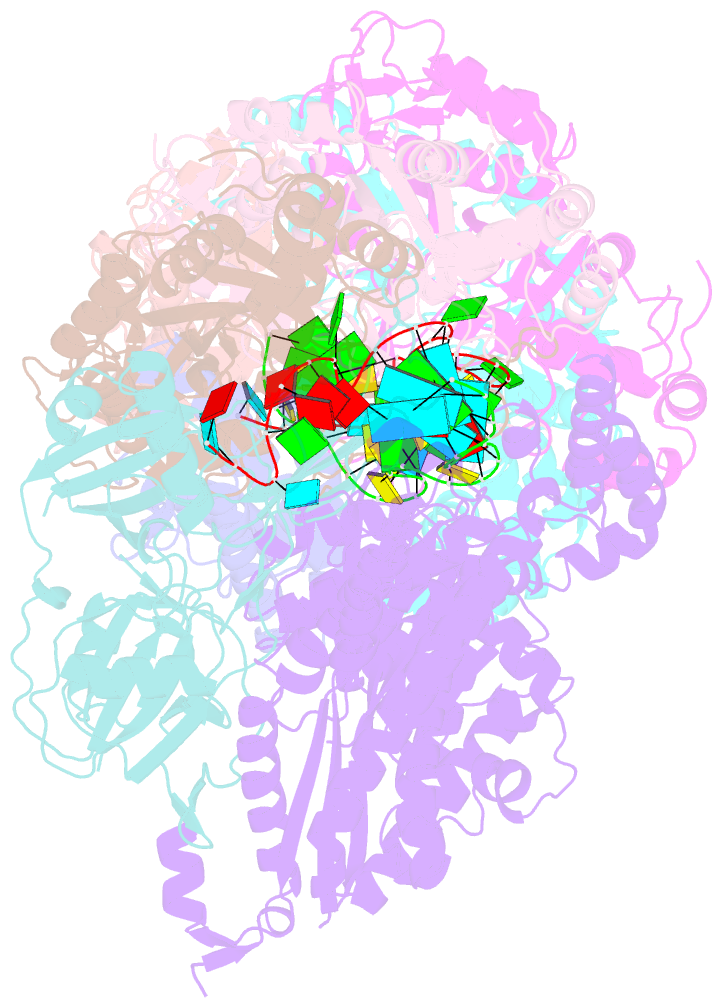
Summary information and primary citation
- PDB-id
- 3x1l; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA-DNA
- Method
- X-ray (2.096 Å)
- Summary
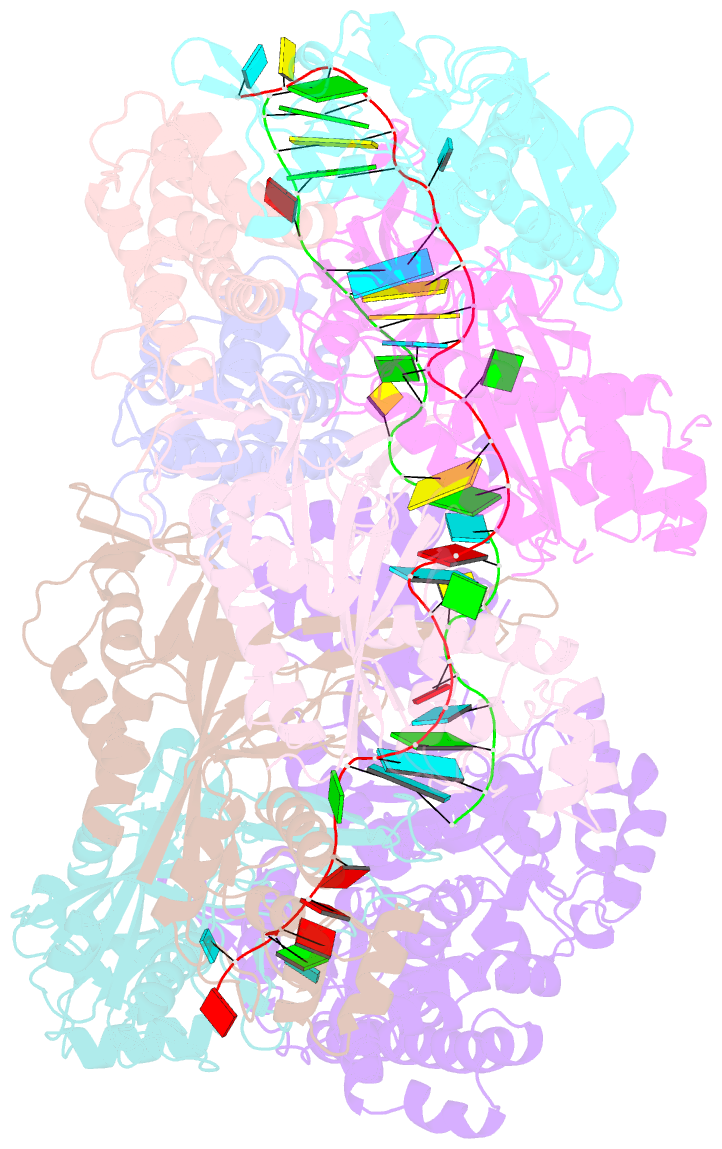
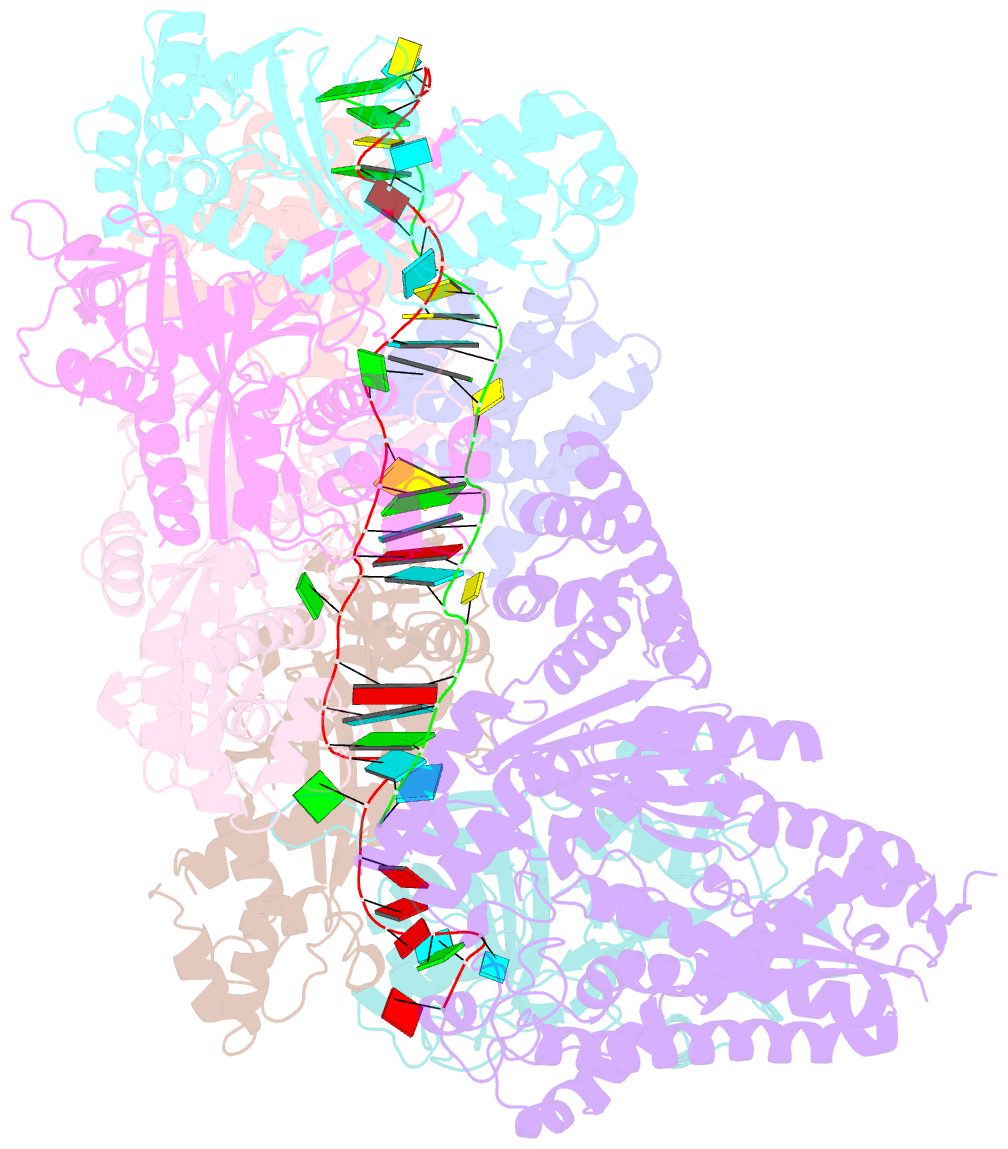
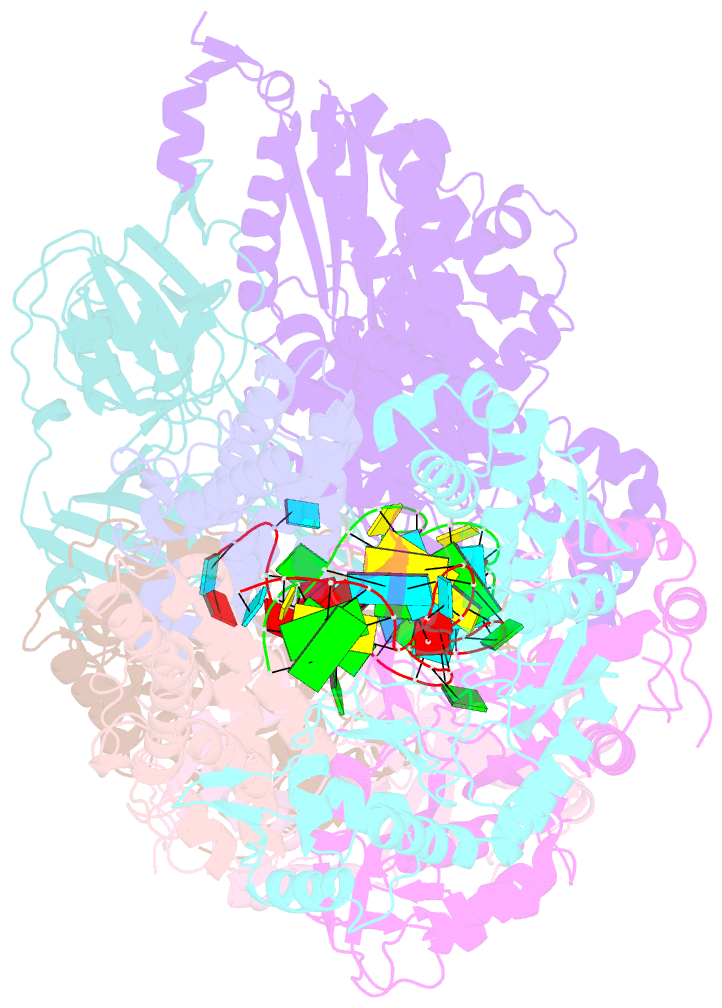
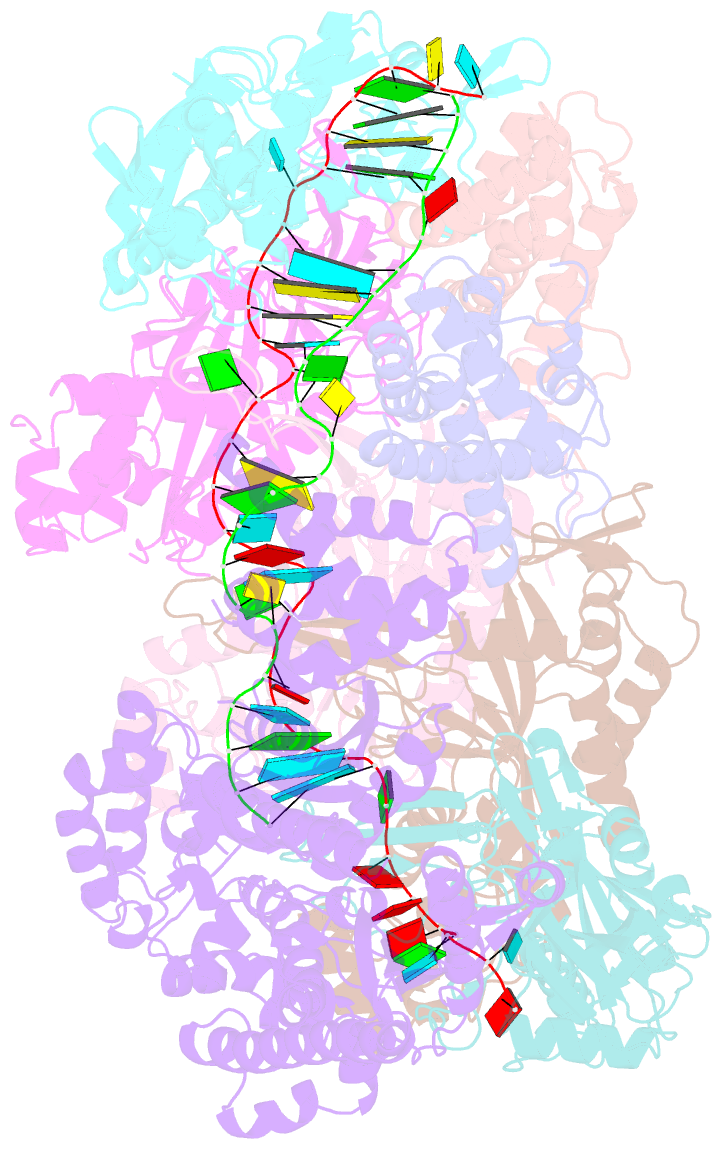
- Crystal structure of the crispr-cas RNA silencing cmr complex bound to a target analog
- Reference
- Osawa T, Inanaga H, Sato C, Numata T (2015): "Crystal structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog." Mol.Cell, 58, 418-430. doi: 10.1016/j.molcel.2015.03.018.
- Abstract
- In prokaryotes, Clustered regularly interspaced short palindromic repeat (CRISPR)-derived RNAs (crRNAs), together with CRISPR-associated (Cas) proteins, capture and degrade invading genetic materials. In the type III-B CRISPR-Cas system, six Cas proteins (Cmr1-Cmr6) and a crRNA form an RNA silencing Cmr complex. Here we report the 2.1 Å crystal structure of the Cmr1-deficient, functional Cmr complex bound to single-stranded DNA, a substrate analog complementary to the crRNA guide. Cmr3 recognizes the crRNA 5' tag and defines the start position of the guide-target duplex, using its idiosyncratic loops. The β-hairpins of three Cmr4 subunits intercalate within the duplex, causing nucleotide displacements with 6 nt intervals, and thus periodically placing the scissile bonds near the crucial aspartate of Cmr4. The structure reveals the mechanism for specifying the periodic target cleavage sites from the crRNA 5' tag and provides insights into the assembly of the type III interference machineries and the evolution of the Cmr and Cascade complexes.