Summary information and primary citation
- PDB-id
- 3zkc; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.0 Å)
- Summary
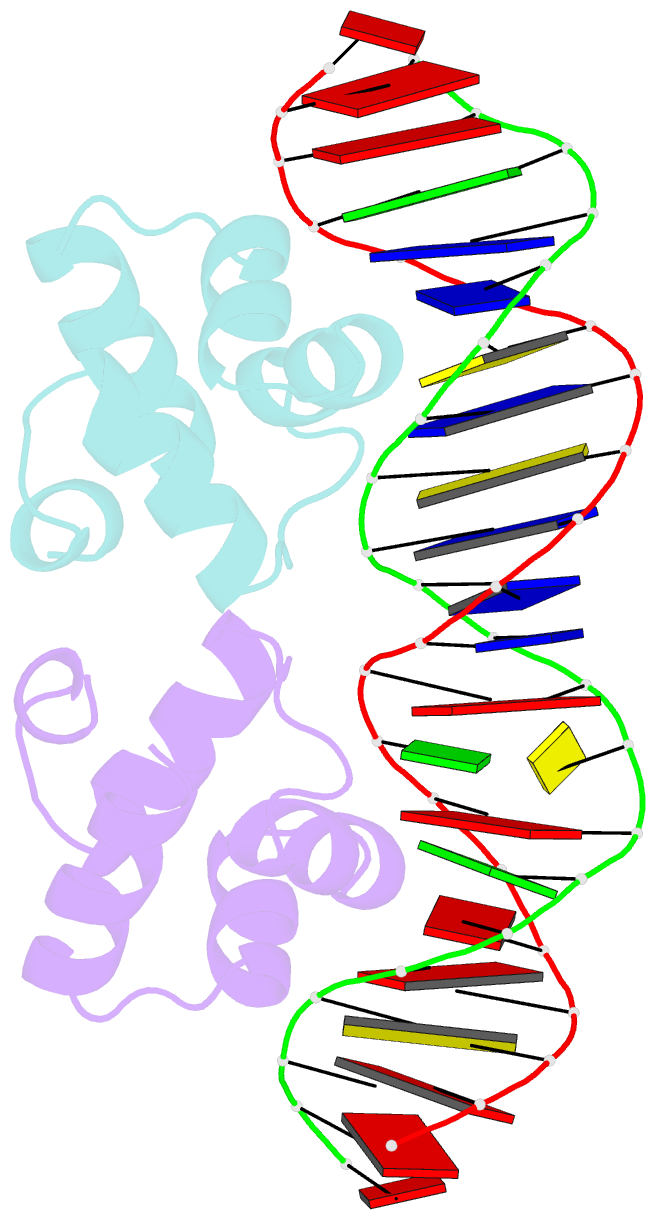
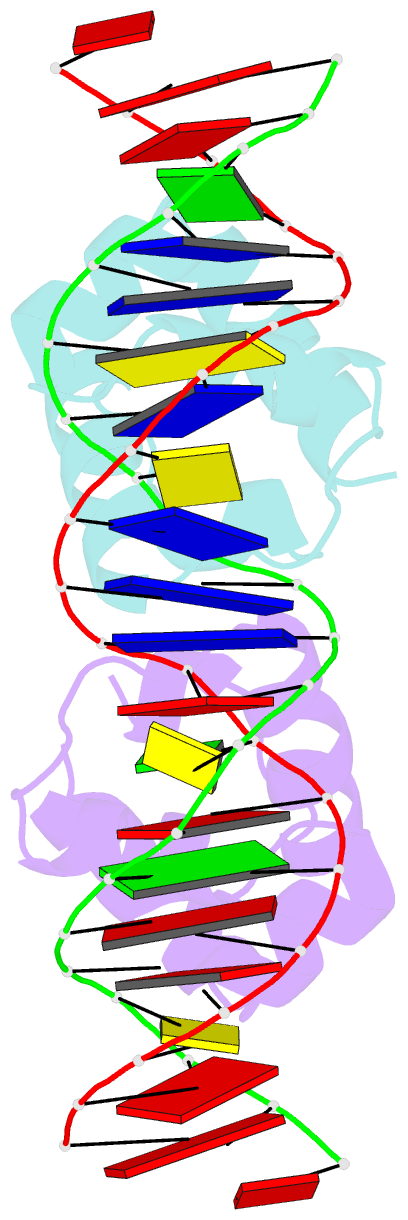
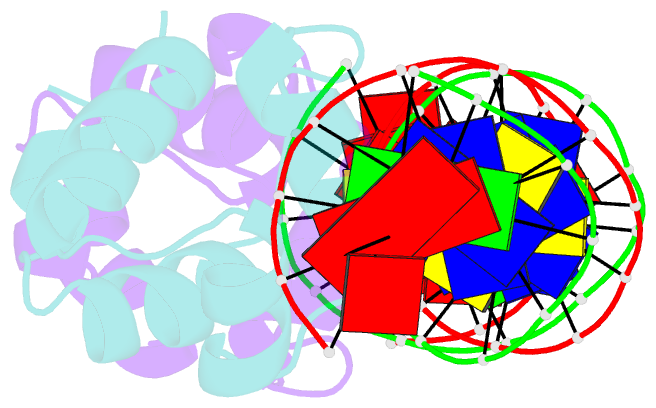
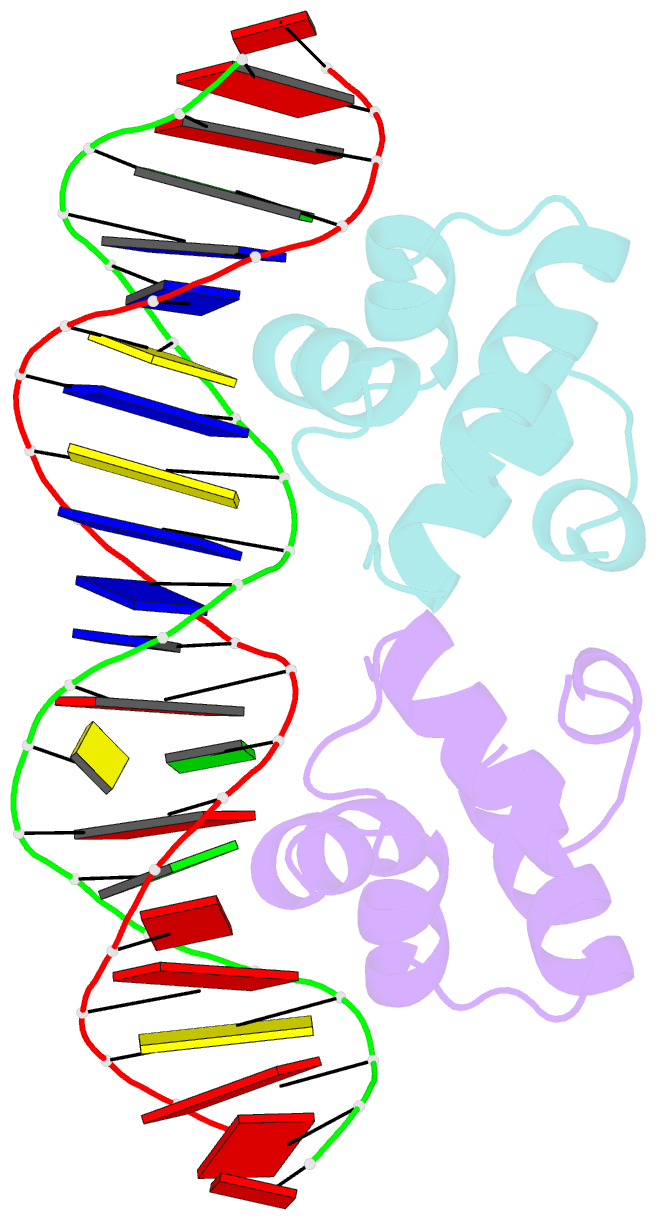
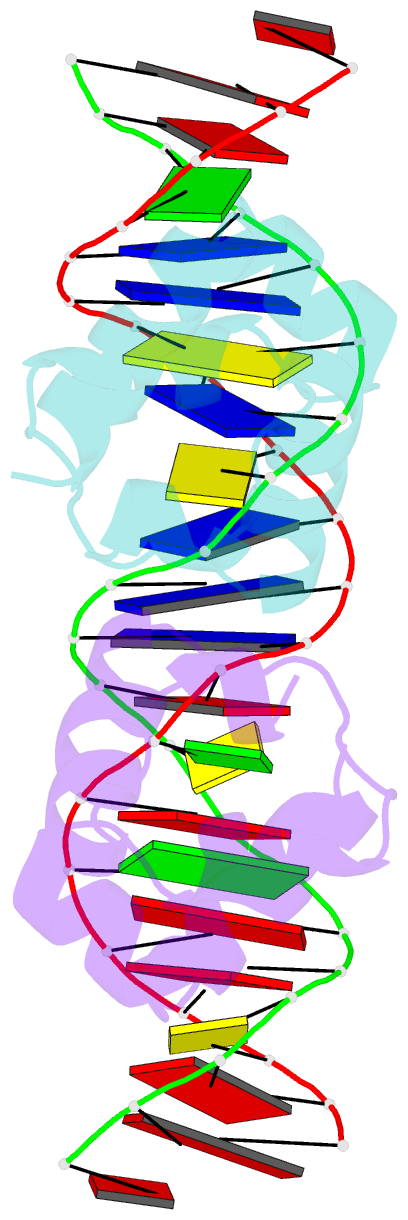
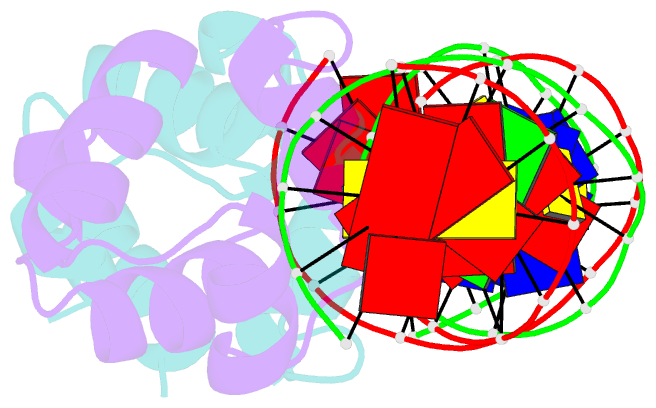
- Crystal structure of the master regulator for biofilm formation sinr in complex with DNA.
- Reference
- Newman JA, Rodrigues C, Lewis RJ (2013): "Molecular Basis of the Activity of Sinr, the Master Regulator of Biofilm Formation in Bacillus Subtilis." J.Biol.Chem., 288, 10766. doi: 10.1074/JBC.M113.455592.
- Abstract
- Bacterial biofilms are complex communities of cells that are attached to a surface by an extracellular matrix. Biofilms are an increasing environmental and healthcare issue, causing problems ranging from the biofouling of ocean-going vessels, to dental plaque, infections of the urinary tract, and contamination of medical instruments such as catheters. A complete understanding of biofilm formation therefore requires knowledge of the regulatory pathways underpinning its formation so that effective intervention strategies can be determined. The master regulator that determines whether the gram-positive model organism Bacillus subtilis switches from a free-living, planktonic lifestyle to form a biofilm is called SinR. The activity of SinR, a transcriptional regulator, is controlled by its antagonists, SinI, SlrA, and SlrR. The interaction of these four proteins forms a switch, which determines whether or not SinR can inhibit biofilm formation by its repression of a number of extracellular matrix-associated operons. To determine the thermodynamic and kinetic parameters governing the protein-protein and protein-DNA interactions at the heart of this epigenetic switch, we have analyzed the protein-protein and protein-DNA interactions by isothermal titration calorimetry and surface plasmon resonance. We also present the crystal structure of SinR in complex with DNA, revealing the molecular basis of base-specific DNA recognition by SinR and suggesting that the most effective means of transcriptional control occurs by the looping of promoter DNA. The structural analysis also enables predictions about how SinR activity is controlled by its interaction with its antagonists.