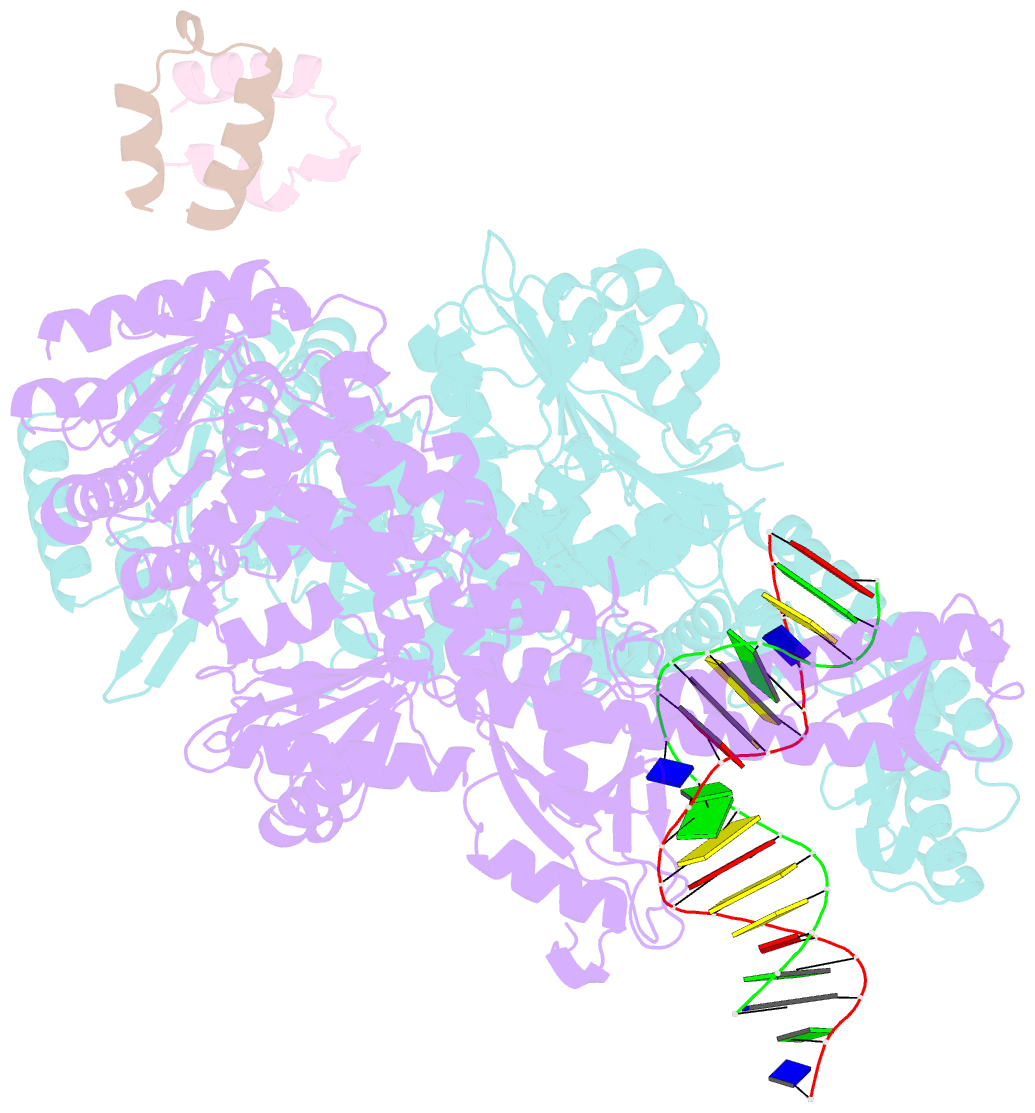
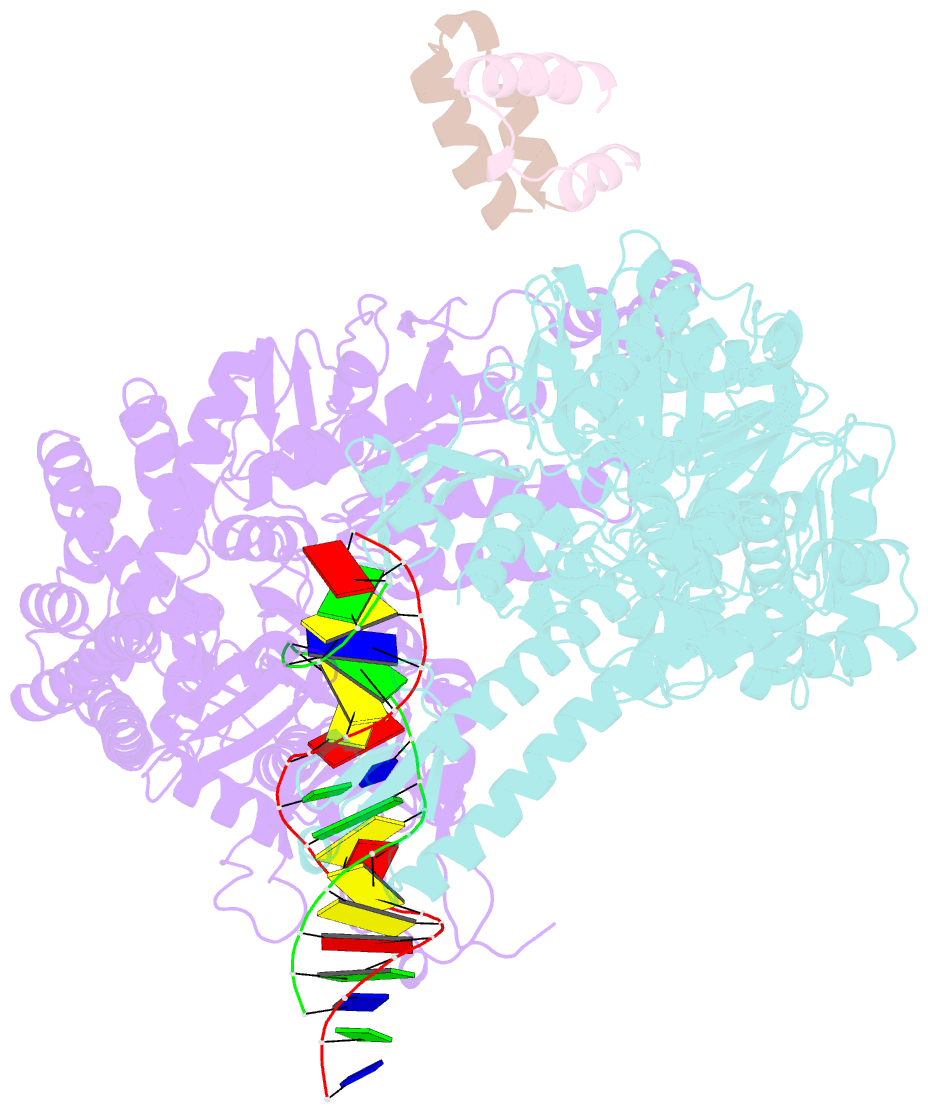
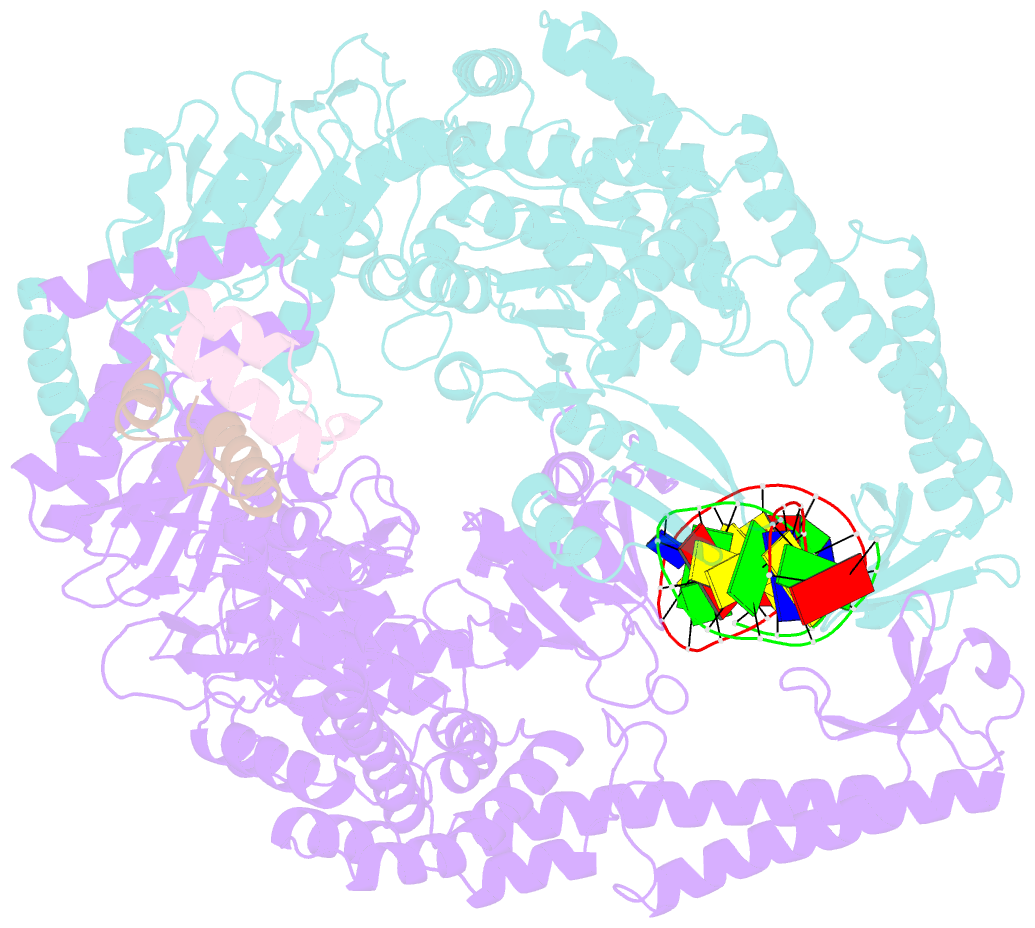
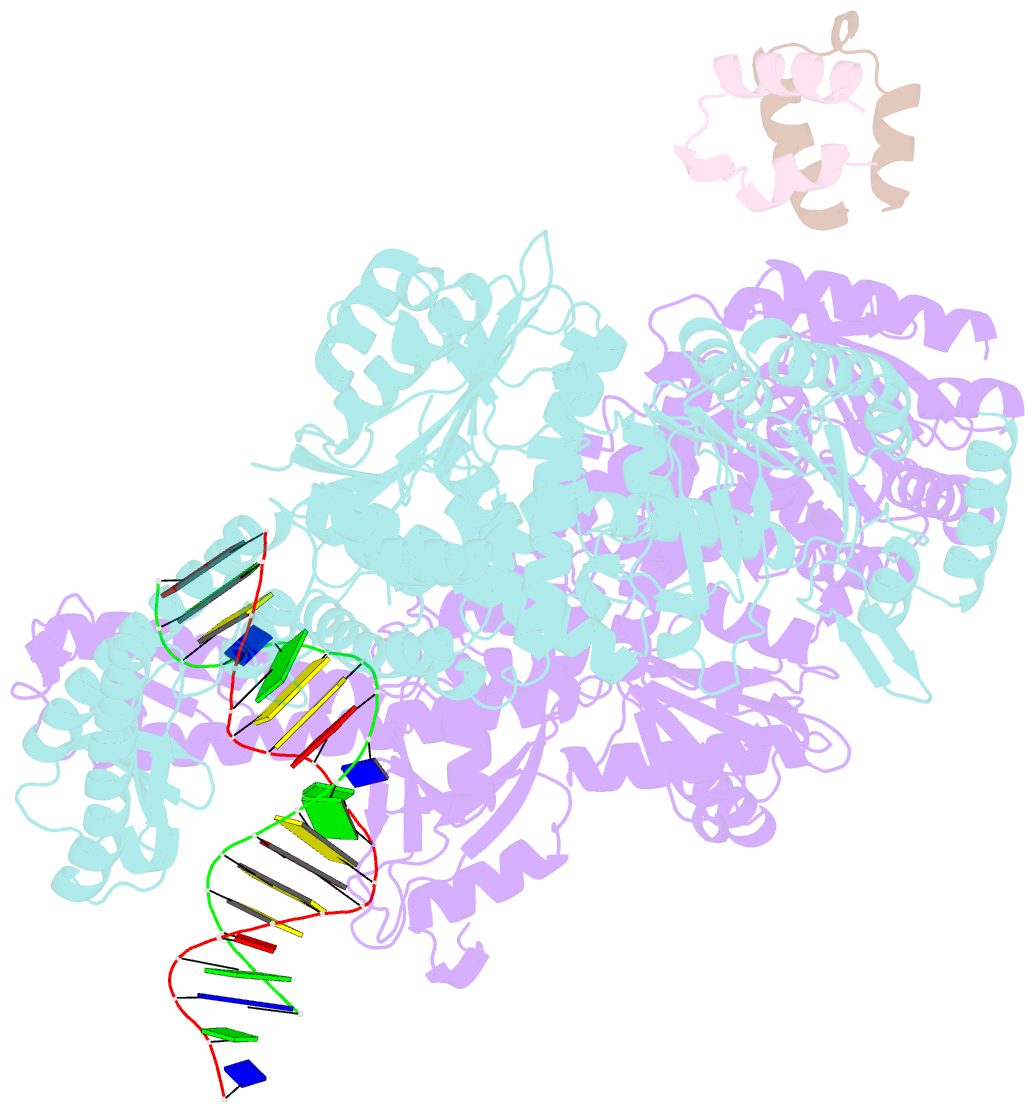
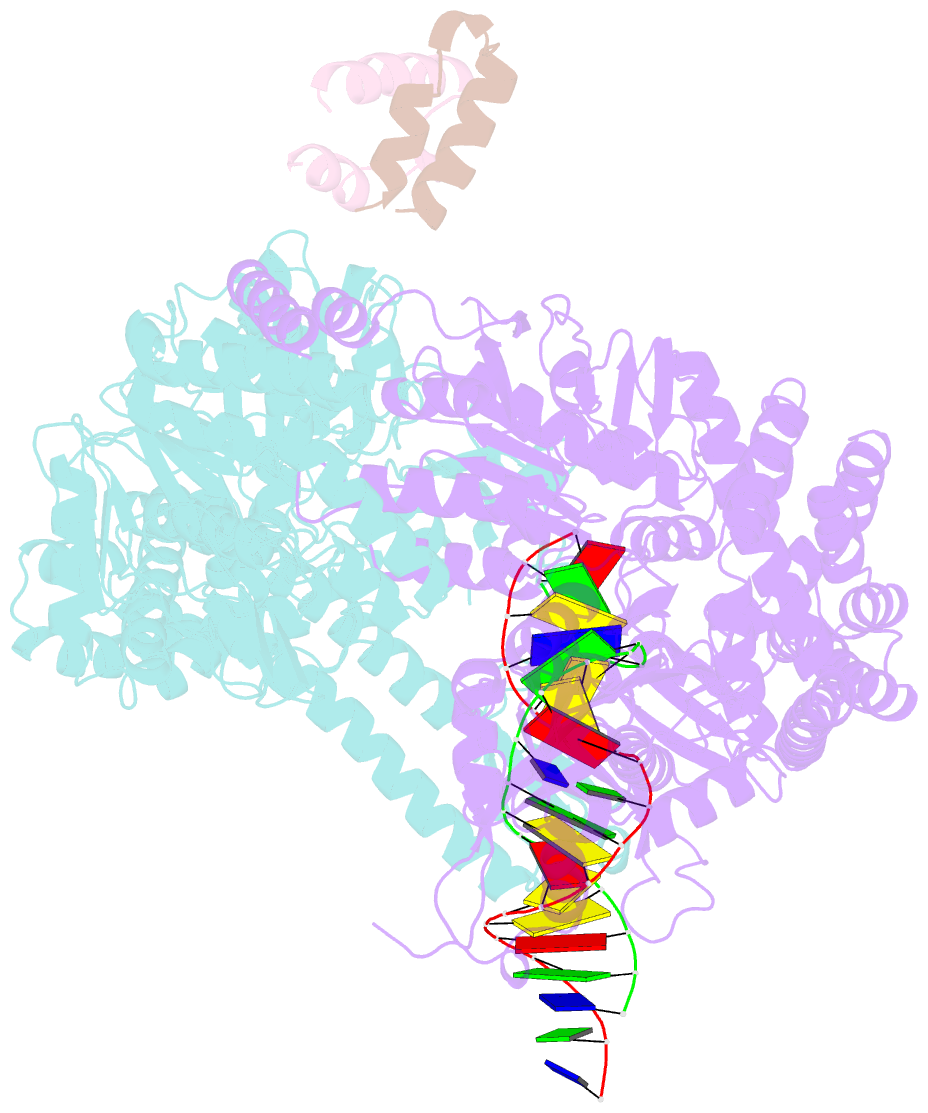
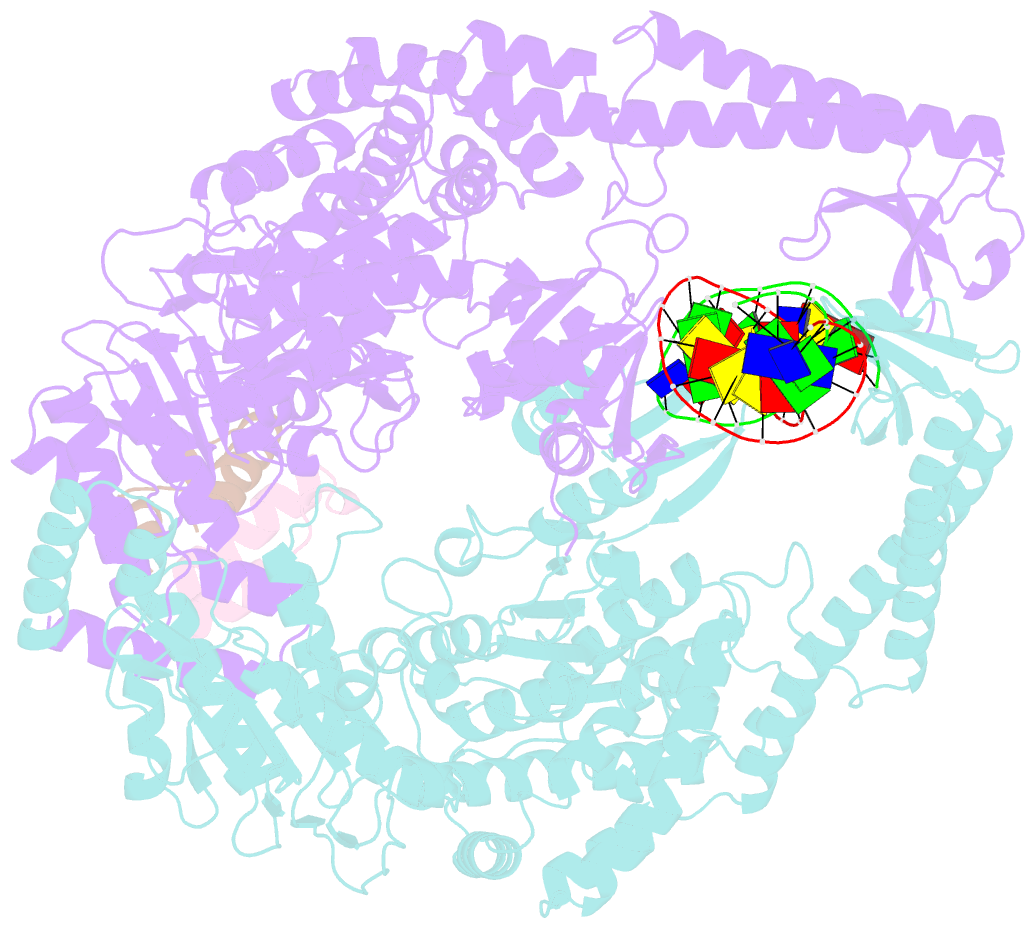
Summary information and primary citation
- PDB-id
- 3zlj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.1 Å)
- Summary
- Crystal structure of full-length e.coli DNA mismatch repair protein muts d835r mutant in complex with gt mismatched DNA
- Reference
- Groothuizen FS, Fish A, Petoukhov MV, Reumer A, Manelyte L, Winterwerp HHK, Marinus MG, Lebbink JHG, Svergun DI, Friedhoff P, Sixma TK (2013): "Using Stable Muts Dimers and Tetramers to Quantitatively Analyze DNA Mismatch Recognition and Sliding Clamp Formation." Nucleic Acids Res., 41, 8166. doi: 10.1093/NAR/GKT582.
- Abstract
- The process of DNA mismatch repair is initiated when MutS recognizes mismatched DNA bases and starts the repair cascade. The Escherichia coli MutS protein exists in an equilibrium between dimers and tetramers, which has compromised biophysical analysis. To uncouple these states, we have generated stable dimers and tetramers, respectively. These proteins allowed kinetic analysis of DNA recognition and structural analysis of the full-length protein by X-ray crystallography and small angle X-ray scattering. Our structural data reveal that the tetramerization domains are flexible with respect to the body of the protein, resulting in mostly extended structures. Tetrameric MutS has a slow dissociation from DNA, which can be due to occasional bending over and binding DNA in its two binding sites. In contrast, the dimer dissociation is faster, primarily dependent on a combination of the type of mismatch and the flanking sequence. In the presence of ATP, we could distinguish two kinetic groups: DNA sequences where MutS forms sliding clamps and those where sliding clamps are not formed efficiently. Interestingly, this inability to undergo a conformational change rather than mismatch affinity is correlated with mismatch repair.